LEF1 Antibody - #DF7570
| Product: | LEF1 Antibody |
| Catalog: | DF7570 |
| Description: | Rabbit polyclonal antibody to LEF1 |
| Application: | WB IHC IF/ICC |
| Reactivity: | Human, Mouse, Rat |
| Prediction: | Pig, Zebrafish, Bovine, Horse, Sheep, Rabbit, Dog, Xenopus |
| Mol.Wt.: | 55 kDa; 44kD(Calculated). |
| Uniprot: | Q9UJU2 |
| RRID: | AB_2841063 |
Related Downloads
Protocols
Product Info
*The optimal dilutions should be determined by the end user.
*Tips:
WB: For western blot detection of denatured protein samples. IHC: For immunohistochemical detection of paraffin sections (IHC-p) or frozen sections (IHC-f) of tissue samples. IF/ICC: For immunofluorescence detection of cell samples. ELISA(peptide): For ELISA detection of antigenic peptide.
Cite Format: Affinity Biosciences Cat# DF7570, RRID:AB_2841063.
Fold/Unfold
DKFZp586H0919; FLJ46390; LEF 1; LEF-1; Lef1; LEF1_HUMAN; Lymphoid enhancer binding factor 1; Lymphoid enhancer-binding factor 1; T cell specific transcription factor 1 alpha; T cell-specific transcription factor 1-alpha; TCF 1 alpha; TCF1 alpha; TCF1-alpha; TCF10; TCF1alpha; TCF7L3; Transcription factor T cell specific 1 alpha;
Immunogens
Detected in thymus. Not detected in normal colon, but highly expressed in colon cancer biopsies and colon cancer cell lines. Expressed in several pancreatic tumors and weakly expressed in normal pancreatic tissue. Isoforms 1 and 5 are detected in several pancreatic cell lines.
- Q9UJU2 LEF1_HUMAN:
- Protein BLAST With
- NCBI/
- ExPASy/
- Uniprot
MPQLSGGGGGGGGDPELCATDEMIPFKDEGDPQKEKIFAEISHPEEEGDLADIKSSLVNESEIIPASNGHEVARQAQTSQEPYHDKAREHPDDGKHPDGGLYNKGPSYSSYSGYIMMPNMNNDPYMSNGSLSPPIPRTSNKVPVVQPSHAVHPLTPLITYSDEHFSPGSHPSHIPSDVNSKQGMSRHPPAPDIPTFYPLSPGGVGQITPPLGWQGQPVYPITGGFRQPYPSSLSVDTSMSRFSHHMIPGPPGPHTTGIPHPAIVTPQVKQEHPHTDSDLMHVKPQHEQRKEQEPKRPHIKKPLNAFMLYMKEMRANVVAECTLKESAAINQILGRRWHALSREEQAKYYELARKERQLHMQLYPGWSARDNYGKKKKRKREKLQESASGTGPRMTAAYI
Predictions
Score>80(red) has high confidence and is suggested to be used for WB detection. *The prediction model is mainly based on the alignment of immunogen sequences, the results are for reference only, not as the basis of quality assurance.
High(score>80) Medium(80>score>50) Low(score<50) No confidence
PTMs - Q9UJU2 As Substrate
| Site | PTM Type | Enzyme | Source |
|---|---|---|---|
| S5 | Phosphorylation | Uniprot | |
| K36 | Ubiquitination | Uniprot | |
| S42 | Phosphorylation | P68400 (CSNK2A1) | Uniprot |
| K54 | Ubiquitination | Uniprot | |
| S61 | Phosphorylation | P68400 (CSNK2A1) | Uniprot |
| S79 | Phosphorylation | Uniprot | |
| Y83 | Phosphorylation | Uniprot | |
| K86 | Acetylation | Uniprot | |
| K86 | Ubiquitination | Uniprot | |
| K95 | Acetylation | Uniprot | |
| K95 | Ubiquitination | Uniprot | |
| Y102 | Phosphorylation | Uniprot | |
| Y114 | Phosphorylation | Uniprot | |
| S132 | Phosphorylation | Q9UBE8 (NLK) | Uniprot |
| T155 | Phosphorylation | Q9UBE8 (NLK) | Uniprot |
| T159 | Phosphorylation | Uniprot | |
| Y160 | Phosphorylation | Uniprot | |
| S161 | Phosphorylation | Uniprot | |
| S166 | Phosphorylation | Q9UBE8 (NLK) | Uniprot |
| T195 | Phosphorylation | Uniprot | |
| Y197 | Phosphorylation | Uniprot | |
| S200 | Phosphorylation | Q9UBE8 (NLK) | Uniprot |
| T208 | Phosphorylation | Uniprot | |
| R226 | Methylation | Uniprot | |
| S238 | Phosphorylation | Uniprot | |
| S240 | Phosphorylation | Uniprot | |
| R241 | Methylation | Uniprot | |
| T265 | Phosphorylation | Q9UBE8 (NLK) | Uniprot |
| K283 | Ubiquitination | Uniprot | |
| K295 | Ubiquitination | Uniprot | |
| K301 | Ubiquitination | Uniprot | |
| K311 | Ubiquitination | Uniprot | |
| K324 | Ubiquitination | Uniprot | |
| K347 | Ubiquitination | Uniprot | |
| Y348 | Phosphorylation | Uniprot | |
| Y349 | Phosphorylation | Uniprot | |
| Y363 | Phosphorylation | Uniprot | |
| S367 | Phosphorylation | Uniprot | |
| K382 | Ubiquitination | Uniprot | |
| S386 | Phosphorylation | Uniprot | |
| T390 | Phosphorylation | Uniprot | |
| R393 | Methylation | Uniprot |
Research Backgrounds
Participates in the Wnt signaling pathway. Activates transcription of target genes in the presence of CTNNB1 and EP300. May play a role in hair cell differentiation and follicle morphogenesis. TLE1, TLE2, TLE3 and TLE4 repress transactivation mediated by LEF1 and CTNNB1. Regulates T-cell receptor alpha enhancer function. Binds DNA in a sequence-specific manner. PIAG antagonizes both Wnt-dependent and Wnt-independent activation by LEF1 (By similarity). Isoform 3 lacks the CTNNB1 interaction domain and may be an antagonist for Wnt signaling. Isoform 5 transcriptionally activates the fibronectin promoter, binds to and represses transcription from the E-cadherin promoter in a CTNNB1-independent manner, and is involved in reducing cellular aggregation and increasing cell migration of pancreatic cancer cells. Isoform 1 transcriptionally activates MYC and CCND1 expression and enhances proliferation of pancreatic tumor cells.
Phosphorylated at Thr-155 and/or Ser-166 by NLK. Phosphorylation by NLK at these sites represses LEF1-mediated transcriptional activation of target genes of the canonical Wnt signaling pathway.
Nucleus.
Note: Found in nuclear bodies upon PIASG binding.
Detected in thymus. Not detected in normal colon, but highly expressed in colon cancer biopsies and colon cancer cell lines. Expressed in several pancreatic tumors and weakly expressed in normal pancreatic tissue. Isoforms 1 and 5 are detected in several pancreatic cell lines.
Binds the armadillo repeat of CTNNB1 and forms a stable complex. Interacts with EP300, TLE1 and PIASG (By similarity). Binds ALYREF/THOC4, MDFI and MDFIC. Interacts with NLK.
Proline-rich and acidic regions are implicated in the activation functions of RNA polymerase II transcription factors.
Belongs to the TCF/LEF family.
Research Fields
· Cellular Processes > Cellular community - eukaryotes > Adherens junction. (View pathway)
· Environmental Information Processing > Signal transduction > Wnt signaling pathway. (View pathway)
· Environmental Information Processing > Signal transduction > Hippo signaling pathway. (View pathway)
· Human Diseases > Cancers: Overview > Pathways in cancer. (View pathway)
· Human Diseases > Cancers: Specific types > Colorectal cancer. (View pathway)
· Human Diseases > Cancers: Specific types > Endometrial cancer. (View pathway)
· Human Diseases > Cancers: Specific types > Prostate cancer. (View pathway)
· Human Diseases > Cancers: Specific types > Thyroid cancer. (View pathway)
· Human Diseases > Cancers: Specific types > Basal cell carcinoma. (View pathway)
· Human Diseases > Cancers: Specific types > Acute myeloid leukemia. (View pathway)
· Human Diseases > Cancers: Specific types > Breast cancer. (View pathway)
· Human Diseases > Cancers: Specific types > Hepatocellular carcinoma. (View pathway)
· Human Diseases > Cancers: Specific types > Gastric cancer. (View pathway)
· Human Diseases > Cardiovascular diseases > Arrhythmogenic right ventricular cardiomyopathy (ARVC).
· Organismal Systems > Endocrine system > Melanogenesis.
References
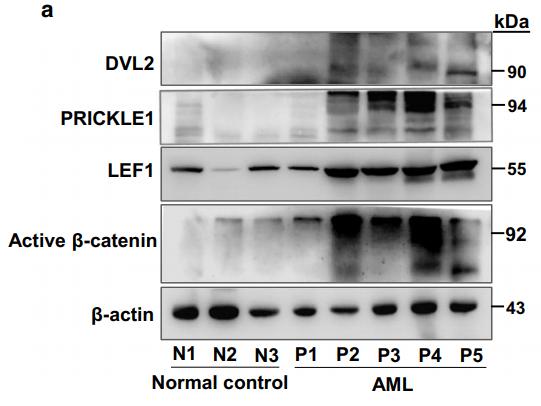
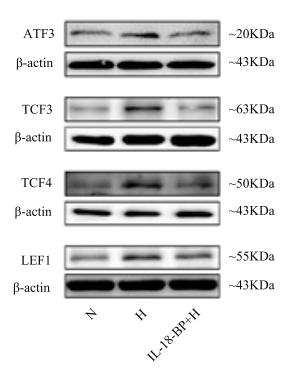
Application: WB Species: human Sample:
Application: WB Species: human Sample: hPDLCs
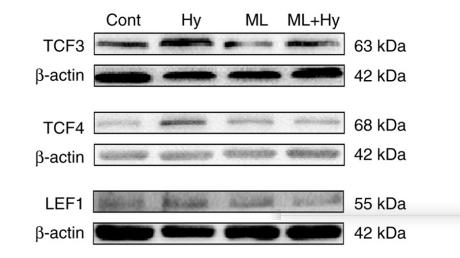
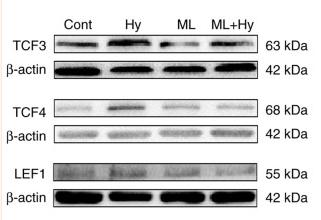
Application: WB Species: rat Sample: atria
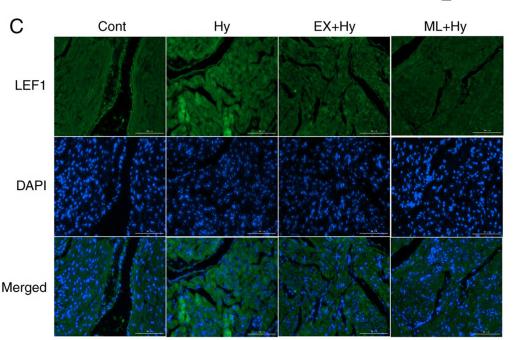
Application: IF/ICC Species: rat Sample: atria
Application: WB Species: Rat Sample:
Application: IF/ICC Species: Rat Sample:
Application: WB Species: rat Sample: atria
Restrictive clause
Affinity Biosciences tests all products strictly. Citations are provided as a resource for additional applications that have not been validated by Affinity Biosciences. Please choose the appropriate format for each application and consult Materials and Methods sections for additional details about the use of any product in these publications.
For Research Use Only.
Not for use in diagnostic or therapeutic procedures. Not for resale. Not for distribution without written consent. Affinity Biosciences will not be held responsible for patent infringement or other violations that may occur with the use of our products. Affinity Biosciences, Affinity Biosciences Logo and all other trademarks are the property of Affinity Biosciences LTD.