Mitofusin 2 Antibody - #DF8106
| Product: | Mitofusin 2 Antibody |
| Catalog: | DF8106 |
| Description: | Rabbit polyclonal antibody to Mitofusin 2 |
| Application: | WB IHC IF/ICC |
| Reactivity: | Human, Mouse, Rat |
| Prediction: | Pig, Zebrafish, Bovine, Horse, Sheep, Rabbit, Dog, Chicken, Xenopus |
| Mol.Wt.: | 86 kDa; 86kD(Calculated). |
| Uniprot: | O95140 |
| RRID: | AB_2841449 |
Related Downloads
Protocols
Product Info
*The optimal dilutions should be determined by the end user.
*Tips:
WB: For western blot detection of denatured protein samples. IHC: For immunohistochemical detection of paraffin sections (IHC-p) or frozen sections (IHC-f) of tissue samples. IF/ICC: For immunofluorescence detection of cell samples. ELISA(peptide): For ELISA detection of antigenic peptide.
Cite Format: Affinity Biosciences Cat# DF8106, RRID:AB_2841449.
Fold/Unfold
CMT2A; CMT2A2; CPRP 1; CPRP1; EC 3.6.5.-; Fzo; HSG; hyperplasia suppressor gene; Hypertension related protein 1; KIAA0214; MARF; MFN 2; Mfn2; MFN2_HUMAN; Mitochondrial assembly regulatory factor; Mitofusin-2; Mitofusin2; Transmembrane GTPase MFN2;
Immunogens
- O95140 MFN2_HUMAN:
- Protein BLAST With
- NCBI/
- ExPASy/
- Uniprot
MSLLFSRCNSIVTVKKNKRHMAEVNASPLKHFVTAKKKINGIFEQLGAYIQESATFLEDTYRNAELDPVTTEEQVLDVKGYLSKVRGISEVLARRHMKVAFFGRTSNGKSTVINAMLWDKVLPSGIGHTTNCFLRVEGTDGHEAFLLTEGSEEKRSAKTVNQLAHALHQDKQLHAGSLVSVMWPNSKCPLLKDDLVLMDSPGIDVTTELDSWIDKFCLDADVFVLVANSESTLMQTEKHFFHKVSERLSRPNIFILNNRWDASASEPEYMEEVRRQHMERCTSFLVDELGVVDRSQAGDRIFFVSAKEVLNARIQKAQGMPEGGGALAEGFQVRMFEFQNFERRFEECISQSAVKTKFEQHTVRAKQIAEAVRLIMDSLHMAAREQQVYCEEMREERQDRLKFIDKQLELLAQDYKLRIKQITEEVERQVSTAMAEEIRRLSVLVDDYQMDFHPSPVVLKVYKNELHRHIEEGLGRNMSDRCSTAITNSLQTMQQDMIDGLKPLLPVSVRSQIDMLVPRQCFSLNYDLNCDKLCADFQEDIEFHFSLGWTMLVNRFLGPKNSRRALMGYNDQVQRPIPLTPANPSMPPLPQGSLTQEEFMVSMVTGLASLTSRTSMGILVVGGVVWKAVGWRLIALSFGLYGLLYVYERLTWTTKAKERAFKRQFVEHASEKLQLVISYTGSNCSHQVQQELSGTFAHLCQQVDVTRENLEQEIAAMNKKIEVLDSLQSKAKLLRNKAGWLDSELNMFTHQYLQPSR
Predictions
Score>80(red) has high confidence and is suggested to be used for WB detection. *The prediction model is mainly based on the alignment of immunogen sequences, the results are for reference only, not as the basis of quality assurance.
High(score>80) Medium(80>score>50) Low(score<50) No confidence
PTMs - O95140 As Substrate
| Site | PTM Type | Enzyme | Source |
|---|---|---|---|
| Ubiquitination | Uniprot | ||
| S10 | Phosphorylation | Uniprot | |
| S27 | Phosphorylation | P45984 (MAPK9) | Uniprot |
| K30 | Ubiquitination | Uniprot | |
| K36 | Acetylation | Uniprot | |
| K36 | Ubiquitination | Uniprot | |
| K79 | Ubiquitination | Uniprot | |
| K84 | Ubiquitination | Uniprot | |
| K98 | Ubiquitination | Uniprot | |
| T111 | Phosphorylation | Uniprot | |
| K154 | Ubiquitination | Uniprot | |
| K158 | Ubiquitination | Uniprot | |
| K171 | Ubiquitination | Uniprot | |
| K192 | Ubiquitination | Uniprot | |
| K243 | Acetylation | Uniprot | |
| K243 | Ubiquitination | Uniprot | |
| K307 | Ubiquitination | Uniprot | |
| K316 | Ubiquitination | Uniprot | |
| K355 | Acetylation | Uniprot | |
| K355 | Ubiquitination | Uniprot | |
| K357 | Ubiquitination | Uniprot | |
| K366 | Ubiquitination | Uniprot | |
| K406 | Ubiquitination | Uniprot | |
| K416 | Ubiquitination | Uniprot | |
| K420 | Ubiquitination | Uniprot | |
| T423 | Phosphorylation | Uniprot | |
| S442 | Phosphorylation | Uniprot | |
| K460 | Ubiquitination | Uniprot | |
| K463 | Ubiquitination | Uniprot | |
| K560 | Ubiquitination | Uniprot | |
| K662 | Ubiquitination | Uniprot | |
| K719 | Ubiquitination | Uniprot | |
| K720 | Ubiquitination | Uniprot | |
| K730 | Ubiquitination | Uniprot | |
| K737 | Ubiquitination | Uniprot | |
| S756 | Phosphorylation | Uniprot |
Research Backgrounds
Mitochondrial outer membrane GTPase that mediates mitochondrial clustering and fusion. Mitochondria are highly dynamic organelles, and their morphology is determined by the equilibrium between mitochondrial fusion and fission events. Overexpression induces the formation of mitochondrial networks. Membrane clustering requires GTPase activity and may involve a major rearrangement of the coiled coil domains (Probable). Plays a central role in mitochondrial metabolism and may be associated with obesity and/or apoptosis processes (By similarity). Plays an important role in the regulation of vascular smooth muscle cell proliferation (By similarity). Involved in the clearance of damaged mitochondria via selective autophagy (mitophagy). Is required for PRKN recruitment to dysfunctional mitochondria. Involved in the control of unfolded protein response (UPR) upon ER stress including activation of apoptosis and autophagy during ER stress (By similarity). Acts as an upstream regulator of EIF2AK3 and suppresses EIF2AK3 activation under basal conditions (By similarity).
Phosphorylated by PINK1.
Ubiquitinated by non-degradative ubiquitin by PRKN, promoting mitochondrial fusion; deubiquitination by USP30 inhibits mitochondrial fusion.
Mitochondrion outer membrane>Multi-pass membrane protein.
Note: Colocalizes with BAX during apoptosis.
Ubiquitous; expressed at low level. Highly expressed in heart and kidney.
Forms homomultimers and heteromultimers with MFN1. Oligomerization is essential for mitochondrion fusion (Probable). Interacts with VAT1 (By similarity). Interacts with STOML2; may form heterooligomers. Interacts (phosphorylated) with PRKN. Interacts with EIF2AK3 (By similarity).
A helix bundle is formed by helices from the N-terminal and the C-terminal part of the protein. The GTPase domain cannot be expressed by itself, without the helix bundle. Rearrangement of the helix bundle and/or of the coiled coil domains may bring membranes from adjacent mitochondria into close contact, and thereby play a role in mitochondrial fusion.
Belongs to the TRAFAC class dynamin-like GTPase superfamily. Dynamin/Fzo/YdjA family. Mitofusin subfamily.
Research Fields
· Organismal Systems > Immune system > NOD-like receptor signaling pathway. (View pathway)
References
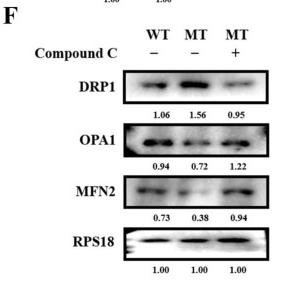
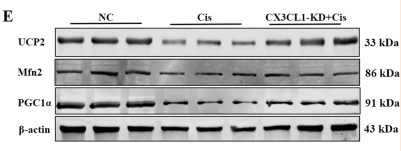
Application: WB Species: Human Sample:
Application: WB Species: Mouse Sample: kidney tissues
Restrictive clause
Affinity Biosciences tests all products strictly. Citations are provided as a resource for additional applications that have not been validated by Affinity Biosciences. Please choose the appropriate format for each application and consult Materials and Methods sections for additional details about the use of any product in these publications.
For Research Use Only.
Not for use in diagnostic or therapeutic procedures. Not for resale. Not for distribution without written consent. Affinity Biosciences will not be held responsible for patent infringement or other violations that may occur with the use of our products. Affinity Biosciences, Affinity Biosciences Logo and all other trademarks are the property of Affinity Biosciences LTD.