Phospho-PPAR gamma (Ser112) Antibody - #AF3284
| Product: | Phospho-PPAR gamma (Ser112) Antibody |
| Catalog: | AF3284 |
| Description: | Rabbit polyclonal antibody to Phospho-PPAR gamma (Ser112) |
| Application: | WB IHC IF/ICC |
| Reactivity: | Human, Mouse, Rat |
| Prediction: | Pig, Bovine, Horse, Sheep, Rabbit, Dog |
| Mol.Wt.: | 57kDa; 58kD(Calculated). |
| Uniprot: | P37231 |
| RRID: | AB_2834705 |
Product Info
*The optimal dilutions should be determined by the end user.
*Tips:
WB: For western blot detection of denatured protein samples. IHC: For immunohistochemical detection of paraffin sections (IHC-p) or frozen sections (IHC-f) of tissue samples. IF/ICC: For immunofluorescence detection of cell samples. ELISA(peptide): For ELISA detection of antigenic peptide.
Cite Format: Affinity Biosciences Cat# AF3284, RRID:AB_2834705.
Fold/Unfold
CIMT1; GLM1; NR1C3; Nuclear receptor subfamily 1 group C member 3; OTTHUMP00000185032; OTTHUMP00000185036; Peroxisome proliferator activated nuclear receptor gamma variant 1; Peroxisome proliferator activated receptor gamma 1; Peroxisome Proliferator Activated Receptor gamma; Peroxisome proliferator-activated receptor gamma; PPAR gamma; PPAR-gamma; PPARG; PPARG_HUMAN; PPARG1; PPARG2; PPARgamma;
Immunogens
Highest expression in adipose tissue. Lower in skeletal muscle, spleen, heart and liver. Also detectable in placenta, lung and ovary.
- P37231 PPARG_HUMAN:
- Protein BLAST With
- NCBI/
- ExPASy/
- Uniprot
MGETLGDSPIDPESDSFTDTLSANISQEMTMVDTEMPFWPTNFGISSVDLSVMEDHSHSFDIKPFTTVDFSSISTPHYEDIPFTRTDPVVADYKYDLKLQEYQSAIKVEPASPPYYSEKTQLYNKPHEEPSNSLMAIECRVCGDKASGFHYGVHACEGCKGFFRRTIRLKLIYDRCDLNCRIHKKSRNKCQYCRFQKCLAVGMSHNAIRFGRMPQAEKEKLLAEISSDIDQLNPESADLRALAKHLYDSYIKSFPLTKAKARAILTGKTTDKSPFVIYDMNSLMMGEDKIKFKHITPLQEQSKEVAIRIFQGCQFRSVEAVQEITEYAKSIPGFVNLDLNDQVTLLKYGVHEIIYTMLASLMNKDGVLISEGQGFMTREFLKSLRKPFGDFMEPKFEFAVKFNALELDDSDLAIFIAVIILSGDRPGLLNVKPIEDIQDNLLQALELQLKLNHPESSQLFAKLLQKMTDLRQIVTEHVQLLQVIKKTETDMSLHPLLQEIYKDLY
Predictions
Score>80(red) has high confidence and is suggested to be used for WB detection. *The prediction model is mainly based on the alignment of immunogen sequences, the results are for reference only, not as the basis of quality assurance.
High(score>80) Medium(80>score>50) Low(score<50) No confidence
PTMs - P37231 As Substrate
| Site | PTM Type | Enzyme | Source |
|---|---|---|---|
| Y78 | Phosphorylation | Uniprot | |
| Y95 | Phosphorylation | Uniprot | |
| Y102 | Phosphorylation | Uniprot | |
| K107 | Sumoylation | Uniprot | |
| S112 | Phosphorylation | P50750 (CDK9) , P45983 (MAPK8) , P50613 (CDK7) , Q02750 (MAP2K1) , P27361 (MAPK3) , P28482 (MAPK1) | Uniprot |
| K184 | Ubiquitination | Uniprot | |
| K185 | Ubiquitination | Uniprot | |
| T269 | Phosphorylation | Uniprot | |
| S273 | Phosphorylation | Q00535 (CDK5) | Uniprot |
| K395 | Sumoylation | Uniprot |
Research Backgrounds
Nuclear receptor that binds peroxisome proliferators such as hypolipidemic drugs and fatty acids. Once activated by a ligand, the nuclear receptor binds to DNA specific PPAR response elements (PPRE) and modulates the transcription of its target genes, such as acyl-CoA oxidase. It therefore controls the peroxisomal beta-oxidation pathway of fatty acids. Key regulator of adipocyte differentiation and glucose homeostasis. ARF6 acts as a key regulator of the tissue-specific adipocyte P2 (aP2) enhancer. Acts as a critical regulator of gut homeostasis by suppressing NF-kappa-B-mediated proinflammatory responses. Plays a role in the regulation of cardiovascular circadian rhythms by regulating the transcription of ARNTL/BMAL1 in the blood vessels (By similarity).
(Microbial infection) Upon treatment with M.tuberculosis or its lipoprotein LpqH, phosphorylation of MAPK p38 and IL-6 production are modulated, probably via this protein.
O-GlcNAcylation at Thr-84 reduces transcriptional activity in adipocytes.
Phosphorylated in basal conditions and dephosphorylated when treated with the ligand. May be dephosphorylated by PPP5C. The phosphorylated form may be inactive and dephosphorylation at Ser-112 induces adipogenic activity (By similarity).
Nucleus. Cytoplasm.
Note: Redistributed from the nucleus to the cytosol through a MAP2K1/MEK1-dependent manner. NOCT enhances its nuclear translocation.
Highest expression in adipose tissue. Lower in skeletal muscle, spleen, heart and liver. Also detectable in placenta, lung and ovary.
Interacts with FOXO1 (acetylated form) (By similarity). Heterodimer with other nuclear receptors, such as RXRA. The heterodimer with the retinoic acid receptor RXRA is called adipocyte-specific transcription factor ARF6. Interacts with NCOA6 coactivator, leading to a strong increase in transcription of target genes. Interacts with coactivator PPARBP, leading to a mild increase in transcription of target genes. Interacts with NOCA7 in a ligand-inducible manner. Interacts with NCOA1 and NCOA2 LXXLL motifs. Interacts with ASXL1, ASXL2, DNTTIP2, FAM120B, MAP2K1/MEK1, NR0B2, PDPK1, PRDM16, PRMT2 and TGFB1I1. Interacts (when activated by agonist) with PPP5C. Interacts with HELZ2 and THRAP3; the interaction stimulates the transcriptional activity of PPARG. Interacts with PER2, the interaction is ligand dependent and blocks PPARG recruitment to target promoters. Interacts with NOCT. Interacts with ACTN4. Interacts (when in the liganded conformation) with GPS2 (By similarity). Interacts with CRY1 and CRY2 in a ligand-dependent manner (By similarity). In the absence of hormonal ligand, interacts with TACC1.
The 9aaTAD motif is a transactivation domain present in a large number of yeast and animal transcription factors.
Belongs to the nuclear hormone receptor family. NR1 subfamily.
Research Fields
· Environmental Information Processing > Signal transduction > AMPK signaling pathway. (View pathway)
· Human Diseases > Neurodegenerative diseases > Huntington's disease.
· Human Diseases > Cancers: Overview > Pathways in cancer. (View pathway)
· Human Diseases > Cancers: Overview > Transcriptional misregulation in cancer.
· Human Diseases > Cancers: Specific types > Thyroid cancer. (View pathway)
· Organismal Systems > Endocrine system > PPAR signaling pathway.
· Organismal Systems > Aging > Longevity regulating pathway. (View pathway)
· Organismal Systems > Development > Osteoclast differentiation. (View pathway)
References
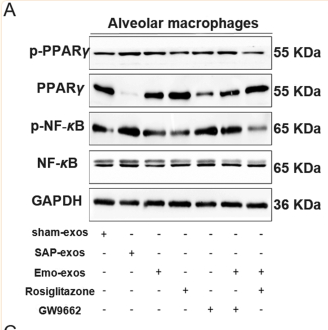
Application: WB Species: Rat Sample: macrophages and lung tissues
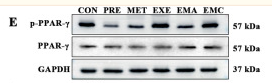
Application: WB Species: rat Sample: dorsal hippocampus
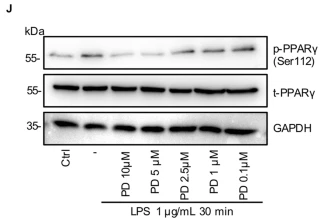
Application: WB Species: Mouse Sample: RAW264.7 cells
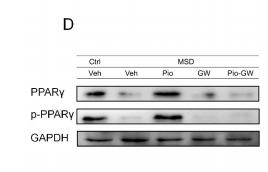
Application: WB Species: Rat Sample: intermuscular adipose tissues (IMATs)
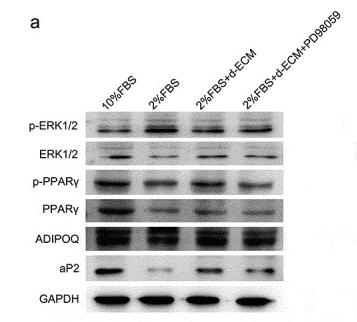
Application: WB Species: Human Sample: ADSCs
Restrictive clause
Affinity Biosciences tests all products strictly. Citations are provided as a resource for additional applications that have not been validated by Affinity Biosciences. Please choose the appropriate format for each application and consult Materials and Methods sections for additional details about the use of any product in these publications.
For Research Use Only.
Not for use in diagnostic or therapeutic procedures. Not for resale. Not for distribution without written consent. Affinity Biosciences will not be held responsible for patent infringement or other violations that may occur with the use of our products. Affinity Biosciences, Affinity Biosciences Logo and all other trademarks are the property of Affinity Biosciences LTD.