NUCL Antibody - #DF12542
| Product: | NUCL Antibody |
| Catalog: | DF12542 |
| Description: | Rabbit polyclonal antibody to NUCL |
| Application: | WB IHC IF/ICC |
| Reactivity: | Human, Mouse, Rat |
| Prediction: | Pig, Sheep, Rabbit, Dog, Xenopus |
| Mol.Wt.: | 110 kDa; 77kD(Calculated). |
| Uniprot: | P19338 |
| RRID: | AB_2845504 |
Related Downloads
Protocols
Product Info
*The optimal dilutions should be determined by the end user.
*Tips:
WB: For western blot detection of denatured protein samples. IHC: For immunohistochemical detection of paraffin sections (IHC-p) or frozen sections (IHC-f) of tissue samples. IF/ICC: For immunofluorescence detection of cell samples. ELISA(peptide): For ELISA detection of antigenic peptide.
Cite Format: Affinity Biosciences Cat# DF12542, RRID:AB_2845504.
Fold/Unfold
C23; FLJ45706; MS1116; NCL; Nucl; NUCL_HUMAN; Nucleolin; Protein C23;
Immunogens
- P19338 NUCL_HUMAN:
- Protein BLAST With
- NCBI/
- ExPASy/
- Uniprot
MVKLAKAGKNQGDPKKMAPPPKEVEEDSEDEEMSEDEEDDSSGEEVVIPQKKGKKAAATSAKKVVVSPTKKVAVATPAKKAAVTPGKKAAATPAKKTVTPAKAVTTPGKKGATPGKALVATPGKKGAAIPAKGAKNGKNAKKEDSDEEEDDDSEEDEEDDEDEDEDEDEIEPAAMKAAAAAPASEDEDDEDDEDDEDDDDDEEDDSEEEAMETTPAKGKKAAKVVPVKAKNVAEDEDEEEDDEDEDDDDDEDDEDDDDEDDEEEEEEEEEEPVKEAPGKRKKEMAKQKAAPEAKKQKVEGTEPTTAFNLFVGNLNFNKSAPELKTGISDVFAKNDLAVVDVRIGMTRKFGYVDFESAEDLEKALELTGLKVFGNEIKLEKPKGKDSKKERDARTLLAKNLPYKVTQDELKEVFEDAAEIRLVSKDGKSKGIAYIEFKTEADAEKTFEEKQGTEIDGRSISLYYTGEKGQNQDYRGGKNSTWSGESKTLVLSNLSYSATEETLQEVFEKATFIKVPQNQNGKSKGYAFIEFASFEDAKEALNSCNKREIEGRAIRLELQGPRGSPNARSQPSKTLFVKGLSEDTTEETLKESFDGSVRARIVTDRETGSSKGFGFVDFNSEEDAKAAKEAMEDGEIDGNKVTLDWAKPKGEGGFGGRGGGRGGFGGRGGGRGGRGGFGGRGRGGFGGRGGFRGGRGGGGDHKPQGKKTKFE
Predictions
Score>80(red) has high confidence and is suggested to be used for WB detection. *The prediction model is mainly based on the alignment of immunogen sequences, the results are for reference only, not as the basis of quality assurance.
High(score>80) Medium(80>score>50) Low(score<50) No confidence
PTMs - P19338 As Substrate
| Site | PTM Type | Enzyme | Source |
|---|---|---|---|
| K3 | Methylation | Uniprot | |
| K9 | Acetylation | Uniprot | |
| K15 | Acetylation | Uniprot | |
| K16 | Acetylation | Uniprot | |
| S28 | Phosphorylation | Uniprot | |
| S34 | Phosphorylation | Uniprot | |
| S41 | Phosphorylation | Uniprot | |
| S42 | Phosphorylation | Uniprot | |
| S60 | Phosphorylation | Uniprot | |
| K62 | Acetylation | Uniprot | |
| K63 | Acetylation | Uniprot | |
| K63 | Ubiquitination | Uniprot | |
| S67 | Phosphorylation | P24941 (CDK2) | Uniprot |
| T69 | Phosphorylation | Uniprot | |
| K70 | Acetylation | Uniprot | |
| K70 | Ubiquitination | Uniprot | |
| K71 | Acetylation | Uniprot | |
| K71 | Ubiquitination | Uniprot | |
| T76 | Phosphorylation | P24941 (CDK2) | Uniprot |
| K79 | Acetylation | Uniprot | |
| K79 | Ubiquitination | Uniprot | |
| K80 | Acetylation | Uniprot | |
| T84 | Phosphorylation | Uniprot | |
| K87 | Acetylation | Uniprot | |
| K88 | Acetylation | Uniprot | |
| T92 | Phosphorylation | Uniprot | |
| K95 | Acetylation | Uniprot | |
| T97 | Phosphorylation | Uniprot | |
| T99 | Phosphorylation | Uniprot | |
| K102 | Acetylation | Uniprot | |
| K102 | Ubiquitination | Uniprot | |
| T105 | Phosphorylation | Uniprot | |
| T106 | Phosphorylation | Uniprot | |
| K109 | Acetylation | Uniprot | |
| K110 | Acetylation | Uniprot | |
| K110 | Ubiquitination | Uniprot | |
| T113 | Phosphorylation | Uniprot | |
| K116 | Acetylation | Uniprot | |
| K116 | Methylation | Uniprot | |
| K116 | Ubiquitination | Uniprot | |
| T121 | Phosphorylation | P06493 (CDK1) , P24941 (CDK2) | Uniprot |
| K124 | Acetylation | Uniprot | |
| K124 | Ubiquitination | Uniprot | |
| K125 | Acetylation | Uniprot | |
| K132 | Acetylation | Uniprot | |
| K135 | Acetylation | Uniprot | |
| S145 | Phosphorylation | Uniprot | |
| S153 | Phosphorylation | Uniprot | |
| S184 | Phosphorylation | Uniprot | |
| S206 | Phosphorylation | Uniprot | |
| T213 | Phosphorylation | Uniprot | |
| T214 | Phosphorylation | Uniprot | |
| K223 | Acetylation | Uniprot | |
| K223 | Sumoylation | Uniprot | |
| K223 | Ubiquitination | Uniprot | |
| K294 | Acetylation | Uniprot | |
| K294 | Sumoylation | Uniprot | |
| K297 | Acetylation | Uniprot | |
| K297 | Sumoylation | Uniprot | |
| T301 | Phosphorylation | Uniprot | |
| T304 | Phosphorylation | Uniprot | |
| T305 | Phosphorylation | Uniprot | |
| K318 | Acetylation | Uniprot | |
| K318 | Ubiquitination | Uniprot | |
| K324 | Sumoylation | Uniprot | |
| K324 | Ubiquitination | Uniprot | |
| T325 | Phosphorylation | Uniprot | |
| S328 | Phosphorylation | Uniprot | |
| K333 | Acetylation | Uniprot | |
| K333 | Methylation | Uniprot | |
| K333 | Ubiquitination | Uniprot | |
| R342 | Methylation | Uniprot | |
| K348 | Ubiquitination | Uniprot | |
| Y351 | Phosphorylation | Uniprot | |
| S356 | Phosphorylation | Uniprot | |
| K362 | Ubiquitination | Uniprot | |
| T367 | Phosphorylation | Uniprot | |
| K370 | Acetylation | Uniprot | |
| K370 | Ubiquitination | Uniprot | |
| K377 | Acetylation | Uniprot | |
| K377 | Sumoylation | Uniprot | |
| K377 | Ubiquitination | Uniprot | |
| K382 | Ubiquitination | Uniprot | |
| K387 | Acetylation | Uniprot | |
| K388 | Acetylation | Uniprot | |
| K398 | Acetylation | Uniprot | |
| K398 | Ubiquitination | Uniprot | |
| Y402 | Phosphorylation | Uniprot | |
| K403 | Acetylation | Uniprot | |
| K403 | Sumoylation | Uniprot | |
| K403 | Ubiquitination | Uniprot | |
| T405 | Phosphorylation | Uniprot | |
| K410 | Ubiquitination | Uniprot | |
| R420 | Methylation | Uniprot | |
| S423 | Phosphorylation | Uniprot | |
| S428 | Phosphorylation | Uniprot | |
| K429 | Ubiquitination | Uniprot | |
| Y433 | Phosphorylation | Uniprot | |
| K437 | Acetylation | Uniprot | |
| K444 | Acetylation | Uniprot | |
| K444 | Ubiquitination | Uniprot | |
| K449 | Acetylation | Uniprot | |
| K449 | Ubiquitination | Uniprot | |
| S458 | Phosphorylation | Uniprot | |
| S460 | Phosphorylation | Uniprot | |
| Y462 | Phosphorylation | Uniprot | |
| Y463 | Phosphorylation | Uniprot | |
| T464 | Phosphorylation | Uniprot | |
| K467 | Acetylation | Uniprot | |
| K467 | Ubiquitination | Uniprot | |
| Y473 | Phosphorylation | Uniprot | |
| R474 | Methylation | Uniprot | |
| K477 | Acetylation | Uniprot | |
| K477 | Ubiquitination | Uniprot | |
| T480 | Phosphorylation | Uniprot | |
| S482 | Phosphorylation | Uniprot | |
| K486 | Ubiquitination | Uniprot | |
| T487 | Phosphorylation | Uniprot | |
| S494 | Phosphorylation | Uniprot | |
| S496 | Phosphorylation | Uniprot | |
| T498 | Phosphorylation | Uniprot | |
| K508 | Ubiquitination | Uniprot | |
| K513 | Acetylation | Uniprot | |
| K513 | Ubiquitination | Uniprot | |
| K521 | Ubiquitination | Uniprot | |
| K523 | Ubiquitination | Uniprot | |
| Y525 | Phosphorylation | Uniprot | |
| S532 | Phosphorylation | Uniprot | |
| K537 | Ubiquitination | Uniprot | |
| S542 | Phosphorylation | Uniprot | |
| C543 | S-Nitrosylation | Uniprot | |
| K545 | Acetylation | Uniprot | |
| K545 | Ubiquitination | Uniprot | |
| R561 | Methylation | Uniprot | |
| S563 | Phosphorylation | Uniprot | |
| K572 | Acetylation | Uniprot | |
| K572 | Ubiquitination | Uniprot | |
| K577 | Acetylation | Uniprot | |
| K577 | Sumoylation | Uniprot | |
| K577 | Ubiquitination | Uniprot | |
| S580 | Phosphorylation | Uniprot | |
| T583 | Phosphorylation | Uniprot | |
| T587 | Phosphorylation | Uniprot | |
| K589 | Sumoylation | Uniprot | |
| K589 | Ubiquitination | Uniprot | |
| S591 | Phosphorylation | Uniprot | |
| S595 | Phosphorylation | Uniprot | |
| R597 | Methylation | Uniprot | |
| T602 | Phosphorylation | Uniprot | |
| R604 | Methylation | Uniprot | |
| T606 | Phosphorylation | Uniprot | |
| S608 | Phosphorylation | Uniprot | |
| S609 | Phosphorylation | Uniprot | |
| K610 | Acetylation | Uniprot | |
| K610 | Ubiquitination | Uniprot | |
| S619 | Phosphorylation | Uniprot | |
| K624 | Acetylation | Uniprot | |
| K624 | Ubiquitination | Uniprot | |
| K639 | Ubiquitination | Uniprot | |
| T641 | Phosphorylation | P06493 (CDK1) | Uniprot |
| K646 | Acetylation | Uniprot | |
| K646 | Methylation | Uniprot | |
| K646 | Ubiquitination | Uniprot | |
| R656 | Methylation | Uniprot | |
| R660 | Methylation | Uniprot | |
| R666 | Methylation | Uniprot | |
| R670 | Methylation | Uniprot | |
| R673 | Methylation | Uniprot | |
| R679 | Methylation | Uniprot | |
| R681 | Methylation | Uniprot | |
| R687 | Methylation | Uniprot | |
| R691 | Methylation | Uniprot | |
| R694 | Methylation | Uniprot | |
| T707 | Phosphorylation | P06493 (CDK1) | Uniprot |
Research Backgrounds
Nucleolin is the major nucleolar protein of growing eukaryotic cells. It is found associated with intranucleolar chromatin and pre-ribosomal particles. It induces chromatin decondensation by binding to histone H1. It is thought to play a role in pre-rRNA transcription and ribosome assembly. May play a role in the process of transcriptional elongation. Binds RNA oligonucleotides with 5'-UUAGGG-3' repeats more tightly than the telomeric single-stranded DNA 5'-TTAGGG-3' repeats.
Some glutamate residues are glycylated by TTLL8. This modification occurs exclusively on glutamate residues and results in a glycine chain on the gamma-carboxyl group (By similarity).
Symmetrically methylated by PRMT5.
Nucleus>Nucleolus. Cytoplasm.
Note: Localized in cytoplasmic mRNP granules containing untranslated mRNAs.
Identified in a IGF2BP1-dependent mRNP granule complex containing untranslated mRNAs. Component of the SWAP complex that consists of NPM1, NCL/nucleolin, PARP1 and SWAP70 (By similarity). Component of a complex which is at least composed of HTATSF1/Tat-SF1, the P-TEFb complex components CDK9 and CCNT1, RNA polymerase II, SUPT5H, and NCL/nucleolin. Interacts with AICDA (By similarity). Interacts with APTX. Interacts with C1QBP. Interacts with ERBB4. Interacts (via C-terminus) with FMR1 isoform 6 (via N-terminus). Interacts with GZF1; this interaction is important for nucleolar localization of GZF1. Interacts with NSUN2. Interacts with NVL. Interacts (via N-terminus domain) with SETX. Interacts (via RRM1 and C-terminal RRM4/Arg/Gly-rich domains) with TERT; the interaction is important for nucleolar localization of TERT. Interacts with WDR46. Interacts with ZFP36. Interacts with LRRC34 (By similarity). Interacts with RRP1B. Interacts with HNRNPU; this interaction occurs during mitosis. Interacts with RIOK1; RIOK1 recruits NCL to PRMT5 for symmetrically methylation. Interacts with ZBTB7B (By similarity). Interacts with MDK; this interaction promotes NCL clustering and lateral movements of this complex into lipid rafts leading to MDK internalization. Interacts with HDGF (isoform 1).
Research Fields
· Human Diseases > Infectious diseases: Bacterial > Pathogenic Escherichia coli infection.
References
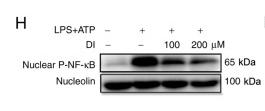
Application: WB Species: Mice Sample: microglial cells
Restrictive clause
Affinity Biosciences tests all products strictly. Citations are provided as a resource for additional applications that have not been validated by Affinity Biosciences. Please choose the appropriate format for each application and consult Materials and Methods sections for additional details about the use of any product in these publications.
For Research Use Only.
Not for use in diagnostic or therapeutic procedures. Not for resale. Not for distribution without written consent. Affinity Biosciences will not be held responsible for patent infringement or other violations that may occur with the use of our products. Affinity Biosciences, Affinity Biosciences Logo and all other trademarks are the property of Affinity Biosciences LTD.