NG2 Antibody - #DF12589
| Product: | NG2 Antibody |
| Catalog: | DF12589 |
| Description: | Rabbit polyclonal antibody to NG2 |
| Application: | WB IHC IF/ICC |
| Reactivity: | Human, Mouse, Rat |
| Prediction: | Pig, Bovine, Horse, Sheep, Rabbit |
| Mol.Wt.: | 251 kDa,320kDa; 251kD(Calculated). |
| Uniprot: | Q6UVK1 |
| RRID: | AB_2845551 |
Related Downloads
Protocols
Product Info
*The optimal dilutions should be determined by the end user.
*Tips:
WB: For western blot detection of denatured protein samples. IHC: For immunohistochemical detection of paraffin sections (IHC-p) or frozen sections (IHC-f) of tissue samples. IF/ICC: For immunofluorescence detection of cell samples. ELISA(peptide): For ELISA detection of antigenic peptide.
Cite Format: Affinity Biosciences Cat# DF12589, RRID:AB_2845551.
Fold/Unfold
4732461B14Rik; AN2; AN2 proteoglycan; Chondroitin sulfate proteoglycan 4 (melanoma-associated); Chondroitin sulfate proteoglycan 4; Chondroitin sulfate proteoglycan NG2; Cspg4; Cspg4 chondroitin sulfate proteoglycan 4; CSPG4_HUMAN; HMW-MAA; HSN tumor-specific antigen; Kiaa4232; MCSP; MCSPG; MEL-CSPG; Melanoma chondroitin sulfate proteoglycan; Melanoma-associated chondroitin sulfate proteoglycan; MELCSPG; MSK16; NG2;
Immunogens
- Q6UVK1 CSPG4_HUMAN:
- Protein BLAST With
- NCBI/
- ExPASy/
- Uniprot
MQSGPRPPLPAPGLALALTLTMLARLASAASFFGENHLEVPVATALTDIDLQLQFSTSQPEALLLLAAGPADHLLLQLYSGRLQVRLVLGQEELRLQTPAETLLSDSIPHTVVLTVVEGWATLSVDGFLNASSAVPGAPLEVPYGLFVGGTGTLGLPYLRGTSRPLRGCLHAATLNGRSLLRPLTPDVHEGCAEEFSASDDVALGFSGPHSLAAFPAWGTQDEGTLEFTLTTQSRQAPLAFQAGGRRGDFIYVDIFEGHLRAVVEKGQGTVLLHNSVPVADGQPHEVSVHINAHRLEISVDQYPTHTSNRGVLSYLEPRGSLLLGGLDAEASRHLQEHRLGLTPEATNASLLGCMEDLSVNGQRRGLREALLTRNMAAGCRLEEEEYEDDAYGHYEAFSTLAPEAWPAMELPEPCVPEPGLPPVFANFTQLLTISPLVVAEGGTAWLEWRHVQPTLDLMEAELRKSQVLFSVTRGARHGELELDIPGAQARKMFTLLDVVNRKARFIHDGSEDTSDQLVLEVSVTARVPMPSCLRRGQTYLLPIQVNPVNDPPHIIFPHGSLMVILEHTQKPLGPEVFQAYDPDSACEGLTFQVLGTSSGLPVERRDQPGEPATEFSCRELEAGSLVYVHRGGPAQDLTFRVSDGLQASPPATLKVVAIRPAIQIHRSTGLRLAQGSAMPILPANLSVETNAVGQDVSVLFRVTGALQFGELQKQGAGGVEGAEWWATQAFHQRDVEQGRVRYLSTDPQHHAYDTVENLALEVQVGQEILSNLSFPVTIQRATVWMLRLEPLHTQNTQQETLTTAHLEATLEEAGPSPPTFHYEVVQAPRKGNLQLQGTRLSDGQGFTQDDIQAGRVTYGATARASEAVEDTFRFRVTAPPYFSPLYTFPIHIGGDPDAPVLTNVLLVVPEGGEGVLSADHLFVKSLNSASYLYEVMERPRHGRLAWRGTQDKTTMVTSFTNEDLLRGRLVYQHDDSETTEDDIPFVATRQGESSGDMAWEEVRGVFRVAIQPVNDHAPVQTISRIFHVARGGRRLLTTDDVAFSDADSGFADAQLVLTRKDLLFGSIVAVDEPTRPIYRFTQEDLRKRRVLFVHSGADRGWIQLQVSDGQHQATALLEVQASEPYLRVANGSSLVVPQGGQGTIDTAVLHLDTNLDIRSGDEVHYHVTAGPRWGQLVRAGQPATAFSQQDLLDGAVLYSHNGSLSPRDTMAFSVEAGPVHTDATLQVTIALEGPLAPLKLVRHKKIYVFQGEAAEIRRDQLEAAQEAVPPADIVFSVKSPPSAGYLVMVSRGALADEPPSLDPVQSFSQEAVDTGRVLYLHSRPEAWSDAFSLDVASGLGAPLEGVLVELEVLPAAIPLEAQNFSVPEGGSLTLAPPLLRVSGPYFPTLLGLSLQVLEPPQHGALQKEDGPQARTLSAFSWRMVEEQLIRYVHDGSETLTDSFVLMANASEMDRQSHPVAFTVTVLPVNDQPPILTTNTGLQMWEGATAPIPAEALRSTDGDSGSEDLVYTIEQPSNGRVVLRGAPGTEVRSFTQAQLDGGLVLFSHRGTLDGGFRFRLSDGEHTSPGHFFRVTAQKQVLLSLKGSQTLTVCPGSVQPLSSQTLRASSSAGTDPQLLLYRVVRGPQLGRLFHAQQDSTGEALVNFTQAEVYAGNILYEHEMPPEPFWEAHDTLELQLSSPPARDVAATLAVAVSFEAACPQRPSHLWKNKGLWVPEGQRARITVAALDASNLLASVPSPQRSEHDVLFQVTQFPSRGQLLVSEEPLHAGQPHFLQSQLAAGQLVYAHGGGGTQQDGFHFRAHLQGPAGASVAGPQTSEAFAITVRDVNERPPQPQASVPLRLTRGSRAPISRAQLSVVDPDSAPGEIEYEVQRAPHNGFLSLVGGGLGPVTRFTQADVDSGRLAFVANGSSVAGIFQLSMSDGASPPLPMSLAVDILPSAIEVQLRAPLEVPQALGRSSLSQQQLRVVSDREEPEAAYRLIQGPQYGHLLVGGRPTSAFSQFQIDQGEVVFAFTNFSSSHDHFRVLALARGVNASAVVNVTVRALLHVWAGGPWPQGATLRLDPTVLDAGELANRTGSVPRFRLLEGPRHGRVVRVPRARTEPGGSQLVEQFTQQDLEDGRLGLEVGRPEGRAPGPAGDSLTLELWAQGVPPAVASLDFATEPYNAARPYSVALLSVPEAARTEAGKPESSTPTGEPGPMASSPEPAVAKGGFLSFLEANMFSVIIPMCLVLLLLALILPLLFYLRKRNKTGKHDVQVLTAKPRNGLAGDTETFRKVEPGQAIPLTAVPGQGPPPGGQPDPELLQFCRTPNPALKNGQYWV
Predictions
Score>80(red) has high confidence and is suggested to be used for WB detection. *The prediction model is mainly based on the alignment of immunogen sequences, the results are for reference only, not as the basis of quality assurance.
High(score>80) Medium(80>score>50) Low(score<50) No confidence
PTMs - Q6UVK1 As Substrate
| Site | PTM Type | Enzyme | Source |
|---|---|---|---|
| T21 | Phosphorylation | Uniprot | |
| S163 | Phosphorylation | Uniprot | |
| S643 | Phosphorylation | Uniprot | |
| Y743 | Phosphorylation | Uniprot | |
| T1591 | Phosphorylation | Uniprot | |
| S2036 | Phosphorylation | Uniprot | |
| T2042 | Phosphorylation | Uniprot | |
| N2075 | N-Glycosylation | Uniprot | |
| T2077 | Phosphorylation | Uniprot | |
| S2079 | Phosphorylation | Uniprot | |
| T2252 | Phosphorylation | P17252 (PRKCA) | Uniprot |
| T2261 | Phosphorylation | Uniprot | |
| K2263 | Ubiquitination | Uniprot | |
| T2274 | Phosphorylation | Uniprot | |
| K2277 | Ubiquitination | Uniprot | |
| T2310 | Phosphorylation | Uniprot | |
| K2316 | Ubiquitination | Uniprot | |
| Y2320 | Phosphorylation | Uniprot |
Research Backgrounds
Proteoglycan playing a role in cell proliferation and migration which stimulates endothelial cells motility during microvascular morphogenesis. May also inhibit neurite outgrowth and growth cone collapse during axon regeneration. Cell surface receptor for collagen alpha 2(VI) which may confer cells ability to migrate on that substrate. Binds through its extracellular N-terminus growth factors, extracellular matrix proteases modulating their activity. May regulate MPP16-dependent degradation and invasion of type I collagen participating in melanoma cells invasion properties. May modulate the plasminogen system by enhancing plasminogen activation and inhibiting angiostatin. Functions also as a signal transducing protein by binding through its cytoplasmic C-terminus scaffolding and signaling proteins. May promote retraction fiber formation and cell polarization through Rho GTPase activation. May stimulate alpha-4, beta-1 integrin-mediated adhesion and spreading by recruiting and activating a signaling cascade through CDC42, ACK1 and BCAR1. May activate FAK and ERK1/ERK2 signaling cascades.
O-glycosylated; contains glycosaminoglycan chondroitin sulfate which are required for proper localization and function in stress fiber formation (By similarity). Involved in interaction with MMP16 and ITGA4.
Phosphorylation by PRKCA regulates its subcellular location and function in cell motility.
Cell membrane>Single-pass type I membrane protein>Extracellular side. Apical cell membrane>Single-pass type I membrane protein>Extracellular side. Cell projection>Lamellipodium membrane>Single-pass type I membrane protein>Extracellular side. Cell surface.
Note: Localized at the apical plasma membrane it relocalizes to the lamellipodia of astrocytoma upon phosphorylation by PRKCA. Localizes to the retraction fibers. Localizes to the plasma membrane of oligodendrocytes (By similarity).
Detected only in malignant melanoma cells.
Interacts with the first PDZ domain of MPDZ. Interacts with PRKCA. Binds TNC, laminin-1, COL5A1 and COL6A2. Interacts with PLG and angiostatin. Binds FGF2 and PDGFA. Interacts with GRIP1, GRIP2 and GRIA2. Forms a ternary complex with GRIP1 and GRIA2 (By similarity). Interacts with LGALS3 and the integrin composed of ITGB1 and ITGA3. Interacts with ITGA4 through its chondroitin sulfate glycosaminoglycan. Interacts with BCAR1, CDC42 and ACK1. Interacts with MMP16.
References
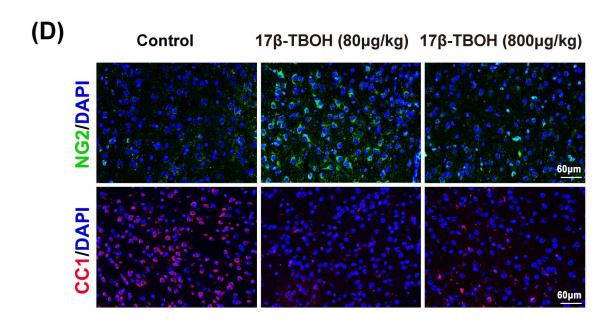
Application: IF/ICC Species: mice Sample: medial prefrontal cortex (mPFC)
Application: WB Species: Mouse Sample:
Application: IF/ICC Species: Mouse Sample:
Application: IF/ICC Species: Mice Sample: retinas
Restrictive clause
Affinity Biosciences tests all products strictly. Citations are provided as a resource for additional applications that have not been validated by Affinity Biosciences. Please choose the appropriate format for each application and consult Materials and Methods sections for additional details about the use of any product in these publications.
For Research Use Only.
Not for use in diagnostic or therapeutic procedures. Not for resale. Not for distribution without written consent. Affinity Biosciences will not be held responsible for patent infringement or other violations that may occur with the use of our products. Affinity Biosciences, Affinity Biosciences Logo and all other trademarks are the property of Affinity Biosciences LTD.