Cryptochrome 2 Antibody - #DF12919
| Product: | Cryptochrome 2 Antibody |
| Catalog: | DF12919 |
| Description: | Rabbit polyclonal antibody to Cryptochrome 2 |
| Application: | WB |
| Reactivity: | Human, Mouse, Rat |
| Prediction: | Pig, Bovine, Horse, Sheep, Rabbit, Dog |
| Mol.Wt.: | 65 kDa; 67kD(Calculated). |
| Uniprot: | Q49AN0 |
| RRID: | AB_2845880 |
Related Downloads
Protocols
Product Info
*The optimal dilutions should be determined by the end user.
*Tips:
WB: For western blot detection of denatured protein samples. IHC: For immunohistochemical detection of paraffin sections (IHC-p) or frozen sections (IHC-f) of tissue samples. IF/ICC: For immunofluorescence detection of cell samples. ELISA(peptide): For ELISA detection of antigenic peptide.
Cite Format: Affinity Biosciences Cat# DF12919, RRID:AB_2845880.
Fold/Unfold
cry2; CRY2_HUMAN; cryptochrome 2 (photolyase like); Cryptochrome 2; Cryptochrome-2; FLJ10332; growth inhibiting protein 37; HCRY2; KIAA0658; PHLL2; Photolyase like;
Immunogens
Expressed in all tissues examined including fetal brain, fibroblasts, heart, brain, placenta, lung, liver, skeletal muscle, kidney, pancreas, spleen, thymus, prostate, testis, ovary, small intestine, colon and leukocytes. Highest levels in heart and skeletal muscle.
- Q49AN0 CRY2_HUMAN:
- Protein BLAST With
- NCBI/
- ExPASy/
- Uniprot
MAATVATAAAVAPAPAPGTDSASSVHWFRKGLRLHDNPALLAAVRGARCVRCVYILDPWFAASSSVGINRWRFLLQSLEDLDTSLRKLNSRLFVVRGQPADVFPRLFKEWGVTRLTFEYDSEPFGKERDAAIMKMAKEAGVEVVTENSHTLYDLDRIIELNGQKPPLTYKRFQAIISRMELPKKPVGLVTSQQMESCRAEIQENHDETYGVPSLEELGFPTEGLGPAVWQGGETEALARLDKHLERKAWVANYERPRMNANSLLASPTGLSPYLRFGCLSCRLFYYRLWDLYKKVKRNSTPPLSLFGQLLWREFFYTAATNNPRFDRMEGNPICIQIPWDRNPEALAKWAEGKTGFPWIDAIMTQLRQEGWIHHLARHAVACFLTRGDLWVSWESGVRVFDELLLDADFSVNAGSWMWLSCSAFFQQFFHCYCPVGFGRRTDPSGDYIRRYLPKLKAFPSRYIYEPWNAPESIQKAAKCIIGVDYPRPIVNHAETSRLNIERMKQIYQQLSRYRGLCLLASVPSCVEDLSHPVAEPSSSQAGSMSSAGPRPLPSGPASPKRKLEAAEEPPGEELSKRARVAELPTPELPSKDA
Predictions
Score>80(red) has high confidence and is suggested to be used for WB detection. *The prediction model is mainly based on the alignment of immunogen sequences, the results are for reference only, not as the basis of quality assurance.
High(score>80) Medium(80>score>50) Low(score<50) No confidence
PTMs - Q49AN0 As Substrate
| Site | PTM Type | Enzyme | Source |
|---|---|---|---|
| Phosphorylation | Uniprot | ||
| Ubiquitination | Uniprot | ||
| K126 | Ubiquitination | Uniprot | |
| K170 | Ubiquitination | Uniprot | |
| K184 | Ubiquitination | Uniprot | |
| T190 | Phosphorylation | Uniprot | |
| S191 | Phosphorylation | Uniprot | |
| S196 | Phosphorylation | Uniprot | |
| K242 | Ubiquitination | Uniprot | |
| K247 | Ubiquitination | Uniprot | |
| S266 | Phosphorylation | P28482 (MAPK1) | Uniprot |
| T300 | Phosphorylation | Uniprot | |
| Y451 | Phosphorylation | Uniprot | |
| K456 | Ubiquitination | Uniprot | |
| Y462 | Phosphorylation | Uniprot | |
| Y464 | Phosphorylation | Uniprot | |
| K475 | Ubiquitination | Uniprot | |
| K478 | Ubiquitination | Uniprot | |
| K504 | Ubiquitination | Uniprot | |
| S511 | Phosphorylation | Uniprot | |
| S558 | Phosphorylation | Q13627 (DYRK1A) | Uniprot |
| S575 | Phosphorylation | Uniprot |
Research Backgrounds
Transcriptional repressor which forms a core component of the circadian clock. The circadian clock, an internal time-keeping system, regulates various physiological processes through the generation of approximately 24 hour circadian rhythms in gene expression, which are translated into rhythms in metabolism and behavior. It is derived from the Latin roots 'circa' (about) and 'diem' (day) and acts as an important regulator of a wide array of physiological functions including metabolism, sleep, body temperature, blood pressure, endocrine, immune, cardiovascular, and renal function. Consists of two major components: the central clock, residing in the suprachiasmatic nucleus (SCN) of the brain, and the peripheral clocks that are present in nearly every tissue and organ system. Both the central and peripheral clocks can be reset by environmental cues, also known as Zeitgebers (German for 'timegivers'). The predominant Zeitgeber for the central clock is light, which is sensed by retina and signals directly to the SCN. The central clock entrains the peripheral clocks through neuronal and hormonal signals, body temperature and feeding-related cues, aligning all clocks with the external light/dark cycle. Circadian rhythms allow an organism to achieve temporal homeostasis with its environment at the molecular level by regulating gene expression to create a peak of protein expression once every 24 hours to control when a particular physiological process is most active with respect to the solar day. Transcription and translation of core clock components (CLOCK, NPAS2, ARNTL/BMAL1, ARNTL2/BMAL2, PER1, PER2, PER3, CRY1 and CRY2) plays a critical role in rhythm generation, whereas delays imposed by post-translational modifications (PTMs) are important for determining the period (tau) of the rhythms (tau refers to the period of a rhythm and is the length, in time, of one complete cycle). A diurnal rhythm is synchronized with the day/night cycle, while the ultradian and infradian rhythms have a period shorter and longer than 24 hours, respectively. Disruptions in the circadian rhythms contribute to the pathology of cardiovascular diseases, cancer, metabolic syndromes and aging. A transcription/translation feedback loop (TTFL) forms the core of the molecular circadian clock mechanism. Transcription factors, CLOCK or NPAS2 and ARNTL/BMAL1 or ARNTL2/BMAL2, form the positive limb of the feedback loop, act in the form of a heterodimer and activate the transcription of core clock genes and clock-controlled genes (involved in key metabolic processes), harboring E-box elements (5'-CACGTG-3') within their promoters. The core clock genes: PER1/2/3 and CRY1/2 which are transcriptional repressors form the negative limb of the feedback loop and interact with the CLOCK|NPAS2-ARNTL/BMAL1|ARNTL2/BMAL2 heterodimer inhibiting its activity and thereby negatively regulating their own expression. This heterodimer also activates nuclear receptors NR1D1/2 and RORA/B/G, which form a second feedback loop and which activate and repress ARNTL/BMAL1 transcription, respectively. CRY1 and CRY2 have redundant functions but also differential and selective contributions at least in defining the pace of the SCN circadian clock and its circadian transcriptional outputs. Less potent transcriptional repressor in cerebellum and liver than CRY1, though less effective in lengthening the period of the SCN oscillator. Seems to play a critical role in tuning SCN circadian period by opposing the action of CRY1. With CRY1, dispensable for circadian rhythm generation but necessary for the development of intercellular networks for rhythm synchrony. May mediate circadian regulation of cAMP signaling and gluconeogenesis by blocking glucagon-mediated increases in intracellular cAMP concentrations and in CREB1 phosphorylation. Besides its role in the maintenance of the circadian clock, is also involved in the regulation of other processes. Plays a key role in glucose and lipid metabolism modulation, in part, through the transcriptional regulation of genes involved in these pathways, such as LEP or ACSL4. Represses glucocorticoid receptor NR3C1/GR-induced transcriptional activity by binding to glucocorticoid response elements (GREs). Represses the CLOCK-ARNTL/BMAL1 induced transcription of BHLHE40/DEC1. Represses the CLOCK-ARNTL/BMAL1 induced transcription of NAMPT (By similarity). Represses PPARD and its target genes in the skeletal muscle and limits exercise capacity (By similarity). Represses the transcriptional activity of NR1I2 (By similarity).
Phosphorylation on Ser-266 by MAPK is important for the inhibition of CLOCK-ARNTL-mediated transcriptional activity. Phosphorylation by CSKNE requires interaction with PER1 or PER2. Phosphorylated in a circadian manner at Ser-554 and Ser-558 in the suprachiasmatic nucleus (SCN) and liver. Phosphorylation at Ser-558 by DYRK1A promotes subsequent phosphorylation at Ser-554 by GSK3-beta: the two-step phosphorylation at the neighboring Ser residues leads to its proteasomal degradation.
Ubiquitinated by the SCF(FBXL3) and SCF(FBXL21) complexes, regulating the balance between degradation and stabilization. The SCF(FBXL3) complex is mainly nuclear and mediates ubiquitination and subsequent degradation of CRY2. In contrast, cytoplasmic SCF(FBXL21) complex-mediated ubiquitination leads to stabilize CRY2 and counteract the activity of the SCF(FBXL3) complex. The SCF(FBXL3) and SCF(FBXL21) complexes probably mediate ubiquitination at different Lys residues. The SCF(FBXL3) complex recognizes and binds CRY2 phosphorylated at Ser-554 and Ser-558. Ubiquitination may be inhibited by PER2. Deubiquitinated by USP7 (By similarity).
Cytoplasm. Nucleus.
Note: Translocated to the nucleus through interaction with other Clock proteins such as PER2 or ARNTL.
Expressed in all tissues examined including fetal brain, fibroblasts, heart, brain, placenta, lung, liver, skeletal muscle, kidney, pancreas, spleen, thymus, prostate, testis, ovary, small intestine, colon and leukocytes. Highest levels in heart and skeletal muscle.
Component of the circadian core oscillator, which includes the CRY proteins, CLOCK or NPAS2, ARNTL/BMAL1 or ARNTL2/BMAL2, CSNK1D and/or CSNK1E, TIMELESS, and the PER proteins (By similarity). Interacts with TIMELESS (By similarity). Interacts directly with PER1, PER2 and PER3; interaction with PER2 inhibits its ubiquitination and vice versa (By similarity). Interacts with CLOCK-ARNTL/BMAL1 (By similarity). Interacts with CLOCK (By similarity). Interacts with ARNTL/BMAL1 (By similarity). Interacts with NFIL3 (By similarity). Interacts with FBXL3. Interacts with FBXL21 (By similarity). FBXL3, PER2 and the cofactor FAD compete for overlapping binding sites (By similarity). FBXL3 cannot bind CRY2 that interacts already with PER2 or that contains bound FAD (By similarity). Interacts with PPP5C (via TPR repeats); the interaction downregulates the PPP5C phosphatase activity on CSNK1E. Interacts with nuclear receptors AR and NR3C1/GR; the interaction is ligand dependent (By similarity). Interacts with PRKDC and CIART (By similarity). Interacts with ISCA1 (in vitro). Interacts with DDB1, USP7 and TARDBP (By similarity). Interacts with HNF4A. Interacts with PPARA (By similarity). Interacts with PPARD (via domain NR LBD) and NR1I2 (via domain NR LBD) in a ligand-dependent manner (By similarity). Interacts with PPARG, NR1I3 and VDR in a ligand-dependent manner (By similarity).
Belongs to the DNA photolyase class-1 family.
Research Fields
· Organismal Systems > Environmental adaptation > Circadian rhythm. (View pathway)
References
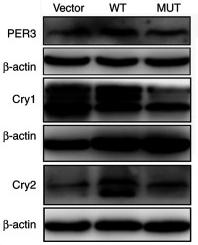
Application: WB Species: Human Sample: U87-MG cells
Restrictive clause
Affinity Biosciences tests all products strictly. Citations are provided as a resource for additional applications that have not been validated by Affinity Biosciences. Please choose the appropriate format for each application and consult Materials and Methods sections for additional details about the use of any product in these publications.
For Research Use Only.
Not for use in diagnostic or therapeutic procedures. Not for resale. Not for distribution without written consent. Affinity Biosciences will not be held responsible for patent infringement or other violations that may occur with the use of our products. Affinity Biosciences, Affinity Biosciences Logo and all other trademarks are the property of Affinity Biosciences LTD.