ROCK1 Antibody - #AF7016
| Product: | ROCK1 Antibody |
| Catalog: | AF7016 |
| Description: | Rabbit polyclonal antibody to ROCK1 |
| Application: | WB IHC IF/ICC |
| Cited expt.: | WB, IHC |
| Reactivity: | Human, Mouse, Rat |
| Prediction: | Pig, Sheep, Rabbit, Dog, Chicken |
| Mol.Wt.: | 160kDa; 158kD(Calculated). |
| Uniprot: | Q13464 |
| RRID: | AB_2835321 |
Product Info
*The optimal dilutions should be determined by the end user. For optimal experimental results, antibody reuse is not recommended.
*Tips:
WB: For western blot detection of denatured protein samples. IHC: For immunohistochemical detection of paraffin sections (IHC-p) or frozen sections (IHC-f) of tissue samples. IF/ICC: For immunofluorescence detection of cell samples. ELISA(peptide): For ELISA detection of antigenic peptide.
Cite Format: Affinity Biosciences Cat# AF7016, RRID:AB_2835321.
Fold/Unfold
coiled-coil-containing protein kinase 1; coiled-coil-containing protein kinase I; MGC131603; MGC43611; p160 Rhoassociated coiled coil-forming protein kinase; p160 ROCK-1; p160 ROCK1; p160ROCK; PRO0435; Renal carcinoma antigen NY REN 35; Renal carcinoma antigen NY-REN-35; Rho associated coiled coil containing protein kinase 1; Rho associated protein kinase 1; Rho kinase; Rho-alpha kinase; Rho-associated; Rho-associated protein kinase 1; ROCK I; ROCK-I; ROCK1; ROCK1_HUMAN; Rok; rokalpha;
Immunogens
A synthesized peptide derived from human ROCK1, corresponding to a region within the internal amino acids.
- Q13464 ROCK1_HUMAN:
- Protein BLAST With
- NCBI/
- ExPASy/
- Uniprot
MSTGDSFETRFEKMDNLLRDPKSEVNSDCLLDGLDALVYDLDFPALRKNKNIDNFLSRYKDTINKIRDLRMKAEDYEVVKVIGRGAFGEVQLVRHKSTRKVYAMKLLSKFEMIKRSDSAFFWEERDIMAFANSPWVVQLFYAFQDDRYLYMVMEYMPGGDLVNLMSNYDVPEKWARFYTAEVVLALDAIHSMGFIHRDVKPDNMLLDKSGHLKLADFGTCMKMNKEGMVRCDTAVGTPDYISPEVLKSQGGDGYYGRECDWWSVGVFLYEMLVGDTPFYADSLVGTYSKIMNHKNSLTFPDDNDISKEAKNLICAFLTDREVRLGRNGVEEIKRHLFFKNDQWAWETLRDTVAPVVPDLSSDIDTSNFDDLEEDKGEEETFPIPKAFVGNQLPFVGFTYYSNRRYLSSANPNDNRTSSNADKSLQESLQKTIYKLEEQLHNEMQLKDEMEQKCRTSNIKLDKIMKELDEEGNQRRNLESTVSQIEKEKMLLQHRINEYQRKAEQENEKRRNVENEVSTLKDQLEDLKKVSQNSQLANEKLSQLQKQLEEANDLLRTESDTAVRLRKSHTEMSKSISQLESLNRELQERNRILENSKSQTDKDYYQLQAILEAERRDRGHDSEMIGDLQARITSLQEEVKHLKHNLEKVEGERKEAQDMLNHSEKEKNNLEIDLNYKLKSLQQRLEQEVNEHKVTKARLTDKHQSIEEAKSVAMCEMEKKLKEEREAREKAENRVVQIEKQCSMLDVDLKQSQQKLEHLTGNKERMEDEVKNLTLQLEQESNKRLLLQNELKTQAFEADNLKGLEKQMKQEINTLLEAKRLLEFELAQLTKQYRGNEGQMRELQDQLEAEQYFSTLYKTQVKELKEEIEEKNRENLKKIQELQNEKETLATQLDLAETKAESEQLARGLLEEQYFELTQESKKAASRNRQEITDKDHTVSRLEEANSMLTKDIEILRRENEELTEKMKKAEEEYKLEKEEEISNLKAAFEKNINTERTLKTQAVNKLAEIMNRKDFKIDRKKANTQDLRKKEKENRKLQLELNQEREKFNQMVVKHQKELNDMQAQLVEECAHRNELQMQLASKESDIEQLRAKLLDLSDSTSVASFPSADETDGNLPESRIEGWLSVPNRGNIKRYGWKKQYVVVSSKKILFYNDEQDKEQSNPSMVLDIDKLFHVRPVTQGDVYRAETEEIPKIFQILYANEGECRKDVEMEPVQQAEKTNFQNHKGHEFIPTLYHFPANCDACAKPLWHVFKPPPALECRRCHVKCHRDHLDKKEDLICPCKVSYDVTSARDMLLLACSQDEQKKWVTHLVKKIPKNPPSGFVRASPRTLSTRSTANQSFRKVVKNTSGKTS
Predictions
Score>80(red) has high confidence and is suggested to be used for WB detection. *The prediction model is mainly based on the alignment of immunogen sequences, the results are for reference only, not as the basis of quality assurance.
High(score>80) Medium(80>score>50) Low(score<50) No confidence
Research Backgrounds
Protein kinase which is a key regulator of actin cytoskeleton and cell polarity. Involved in regulation of smooth muscle contraction, actin cytoskeleton organization, stress fiber and focal adhesion formation, neurite retraction, cell adhesion and motility via phosphorylation of DAPK3, GFAP, LIMK1, LIMK2, MYL9/MLC2, TPPP, PFN1 and PPP1R12A. Phosphorylates FHOD1 and acts synergistically with it to promote SRC-dependent non-apoptotic plasma membrane blebbing. Phosphorylates JIP3 and regulates the recruitment of JNK to JIP3 upon UVB-induced stress. Acts as a suppressor of inflammatory cell migration by regulating PTEN phosphorylation and stability (By similarity). Acts as a negative regulator of VEGF-induced angiogenic endothelial cell activation. Required for centrosome positioning and centrosome-dependent exit from mitosis (By similarity). Plays a role in terminal erythroid differentiation. May regulate closure of the eyelids and ventral body wall by inducing the assembly of actomyosin bundles (By similarity). Promotes keratinocyte terminal differentiation. Involved in osteoblast compaction through the fibronectin fibrillogenesis cell-mediated matrix assembly process, essential for osteoblast mineralization (By similarity).
Autophosphorylated on serine and threonine residues.
Cleaved by caspase-3 during apoptosis. This leads to constitutive activation of the kinase and membrane blebbing.
Cytoplasm. Cytoplasm>Cytoskeleton>Microtubule organizing center>Centrosome>Centriole. Golgi apparatus membrane>Peripheral membrane protein. Cell projection>Bleb. Cytoplasm>Cytoskeleton. Cell membrane. Cell projection>Lamellipodium. Cell projection>Ruffle.
Note: Associated with the mother centriole and an intercentriolar linker. Colocalizes with ITGB1BP1 and ITGB1 at the cell membrane predominantly in lamellipodia and membrane ruffles, but also in retraction fibers. Localizes at the cell membrane in an ITGB1BP1-dependent manner (By similarity). A small proportion is associated with Golgi membranes.
Detected in blood platelets.
The C-terminal auto-inhibitory domain interferes with kinase activity. RHOA binding leads to a conformation change and activation of the kinase. Truncated ROCK1 is constitutively activated.
Belongs to the protein kinase superfamily. AGC Ser/Thr protein kinase family.
Research Fields
· Cellular Processes > Cellular community - eukaryotes > Focal adhesion. (View pathway)
· Cellular Processes > Cellular community - eukaryotes > Tight junction. (View pathway)
· Cellular Processes > Cell motility > Regulation of actin cytoskeleton. (View pathway)
· Environmental Information Processing > Signal transduction > cGMP-PKG signaling pathway. (View pathway)
· Environmental Information Processing > Signal transduction > cAMP signaling pathway. (View pathway)
· Environmental Information Processing > Signal transduction > Sphingolipid signaling pathway. (View pathway)
· Environmental Information Processing > Signal transduction > TGF-beta signaling pathway. (View pathway)
· Human Diseases > Infectious diseases: Bacterial > Pathogenic Escherichia coli infection.
· Human Diseases > Infectious diseases: Bacterial > Shigellosis.
· Human Diseases > Infectious diseases: Bacterial > Salmonella infection.
· Human Diseases > Cancers: Overview > Pathways in cancer. (View pathway)
· Human Diseases > Cancers: Overview > Proteoglycans in cancer.
· Human Diseases > Cancers: Overview > MicroRNAs in cancer.
· Organismal Systems > Immune system > Chemokine signaling pathway. (View pathway)
· Organismal Systems > Circulatory system > Vascular smooth muscle contraction. (View pathway)
· Organismal Systems > Development > Axon guidance. (View pathway)
· Organismal Systems > Immune system > Platelet activation. (View pathway)
· Organismal Systems > Immune system > Leukocyte transendothelial migration. (View pathway)
· Organismal Systems > Endocrine system > Oxytocin signaling pathway.
References
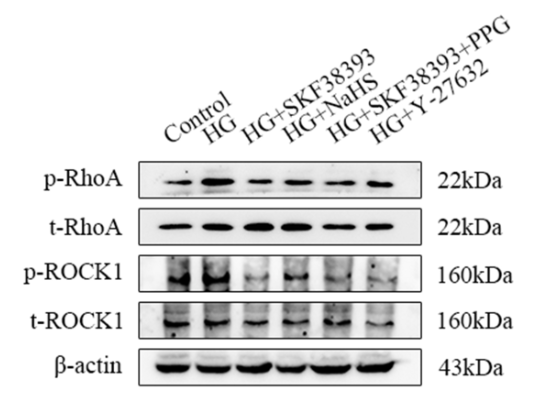
Application: WB Species: mouse Sample: colon
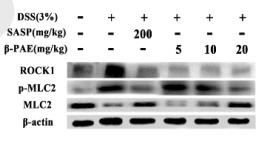
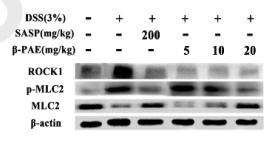
Application: WB Species: mouse Sample: Colons
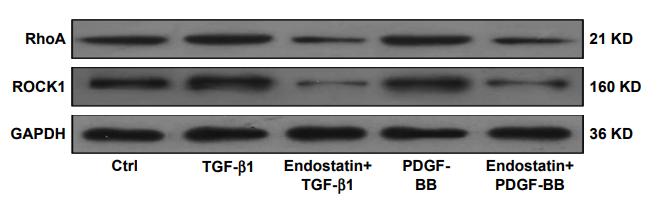
Application: WB Species: rat Sample: HSC-T6 cells
Application: WB Species: Human Sample: 5637 cells
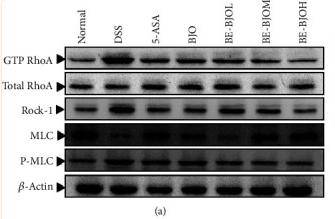
Application: WB Species: Mice Sample: colon tissue
Application: IHC Species: Rat Sample:
Application: WB Species: Human Sample: HUVECs
Restrictive clause
Affinity Biosciences tests all products strictly. Citations are provided as a resource for additional applications that have not been validated by Affinity Biosciences. Please choose the appropriate format for each application and consult Materials and Methods sections for additional details about the use of any product in these publications.
For Research Use Only.
Not for use in diagnostic or therapeutic procedures. Not for resale. Not for distribution without written consent. Affinity Biosciences will not be held responsible for patent infringement or other violations that may occur with the use of our products. Affinity Biosciences, Affinity Biosciences Logo and all other trademarks are the property of Affinity Biosciences LTD.