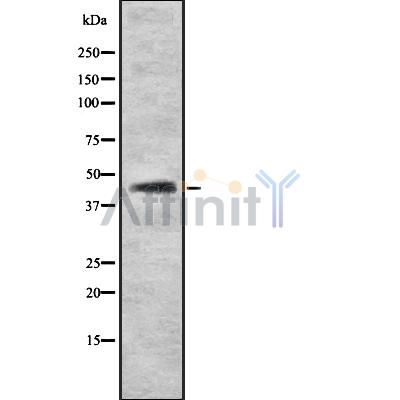
AKAP9 Antibody - #DF8864
| Product: | AKAP9 Antibody |
| Catalog: | DF8864 |
| Description: | Rabbit polyclonal antibody to AKAP9 |
| Application: | WB IHC IF/ICC |
| Reactivity: | Human |
| Prediction: | Pig, Bovine, Sheep, Dog |
| Mol.Wt.: | 454kDa; 453kD(Calculated). |
| Uniprot: | Q99996 |
| RRID: | AB_2842061 |
Related Downloads
Protocols
Product Info
*The optimal dilutions should be determined by the end user. For optimal experimental results, antibody reuse is not recommended.
*Tips:
WB: For western blot detection of denatured protein samples. IHC: For immunohistochemical detection of paraffin sections (IHC-p) or frozen sections (IHC-f) of tissue samples. IF/ICC: For immunofluorescence detection of cell samples. ELISA(peptide): For ELISA detection of antigenic peptide.
Cite Format: Affinity Biosciences Cat# DF8864, RRID:AB_2842061.
Fold/Unfold
A kinase (PRKA) anchor protein (yotiao) 9; A kinase (PRKA) anchor protein 9; A kinase anchor protein 350kDa; A kinase anchor protein 450 kDa; A kinase anchor protein 9; A kinase anchoring protein 350; A kinase anchoring protein 450; A-kinase anchor protein 350 kDa; A-kinase anchor protein 450 kDa; A-kinase anchor protein 9; AKAP 120-like protein; AKAP 350; AKAP 450; AKAP 9; AKAP-9; AKAP120 like protein; AKAP350; AKAP450; AKAP9; AKAP9_HUMAN; Centrosome and Golgi localized PKN associated protein; Centrosome and golgi localized protein; Centrosome- and Golgi-localized PKN-associated protein; CG NAP; CG-NAP; hgAKAP 350; hgAKAP350; Hyperion; KIAA0803; Kinase N associated protein; MU RMS 40.16A; PRKA 9; PRKA9; Protein hyperion; Protein kinase A anchoring protein 9; Protein kinase A-anchoring protein 9; Protein yotiao; Yotiao;
Immunogens
A synthesized peptide derived from human AKAP9, corresponding to a region within C-terminal amino acids.
Widely expressed (PubMed:10202149). Isoform 4: Highly expressed in skeletal muscle and in pancreas (PubMed:9482789).
- Q99996 AKAP9_HUMAN:
- Protein BLAST With
- NCBI/
- ExPASy/
- Uniprot
MEDEERQKKLEAGKAKLAQFRQRKAQSDGQSPSKKQKKKRKTSSSKHDVSAHHDLNIDQSQCNEMYINSSQRVESTVIPESTIMRTLHSGEITSHEQGFSVELESEISTTADDCSSEVNGCSFVMRTGKPTNLLREEEFGVDDSYSEQGAQDSPTHLEMMESELAGKQHEIEELNRELEEMRVTYGTEGLQQLQEFEAAIKQRDGIITQLTANLQQARREKDETMREFLELTEQSQKLQIQFQQLQASETLRNSTHSSTAADLLQAKQQILTHQQQLEEQDHLLEDYQKKKEDFTMQISFLQEKIKVYEMEQDKKVENSNKEEIQEKETIIEELNTKIIEEEKKTLELKDKLTTADKLLGELQEQIVQKNQEIKNMKLELTNSKQKERQSSEEIKQLMGTVEELQKRNHKDSQFETDIVQRMEQETQRKLEQLRAELDEMYGQQIVQMKQELIRQHMAQMEEMKTRHKGEMENALRSYSNITVNEDQIKLMNVAINELNIKLQDTNSQKEKLKEELGLILEEKCALQRQLEDLVEELSFSREQIQRARQTIAEQESKLNEAHKSLSTVEDLKAEIVSASESRKELELKHEAEVTNYKIKLEMLEKEKNAVLDRMAESQEAELERLRTQLLFSHEEELSKLKEDLEIEHRINIEKLKDNLGIHYKQQIDGLQNEMSQKIETMQFEKDNLITKQNQLILEISKLKDLQQSLVNSKSEEMTLQINELQKEIEILRQEEKEKGTLEQEVQELQLKTELLEKQMKEKENDLQEKFAQLEAENSILKDEKKTLEDMLKIHTPVSQEERLIFLDSIKSKSKDSVWEKEIEILIEENEDLKQQCIQLNEEIEKQRNTFSFAEKNFEVNYQELQEEYACLLKVKDDLEDSKNKQELEYKSKLKALNEELHLQRINPTTVKMKSSVFDEDKTFVAETLEMGEVVEKDTTELMEKLEVTKREKLELSQRLSDLSEQLKQKHGEISFLNEEVKSLKQEKEQVSLRCRELEIIINHNRAENVQSCDTQVSSLLDGVVTMTSRGAEGSVSKVNKSFGEESKIMVEDKVSFENMTVGEESKQEQLILDHLPSVTKESSLRATQPSENDKLQKELNVLKSEQNDLRLQMEAQRICLSLVYSTHVDQVREYMENEKDKALCSLKEELIFAQEEKIKELQKIHQLELQTMKTQETGDEGKPLHLLIGKLQKAVSEECSYFLQTLCSVLGEYYTPALKCEVNAEDKENSGDYISENEDPELQDYRYEVQDFQENMHTLLNKVTEEYNKLLVLQTRLSKIWGQQTDGMKLEFGEENLPKEETEFLSIHSQMTNLEDIDVNHKSKLSSLQDLEKTKLEEQVQELESLISSLQQQLKETEQNYEAEIHCLQKRLQAVSESTVPPSLPVDSVVITESDAQRTMYPGSCVKKNIDGTIEFSGEFGVKEETNIVKLLEKQYQEQLEEEVAKVIVSMSIAFAQQTELSRISGGKENTASSKQAHAVCQQEQHYFNEMKLSQDQIGFQTFETVDVKFKEEFKPLSKELGEHGKEILLSNSDPHDIPESKDCVLTISEEMFSKDKTFIVRQSIHDEISVSSMDASRQLMLNEEQLEDMRQELVRQYQEHQQATELLRQAHMRQMERQREDQEQLQEEIKRLNRQLAQRSSIDNENLVSERERVLLEELEALKQLSLAGREKLCCELRNSSTQTQNGNENQGEVEEQTFKEKELDRKPEDVPPEILSNERYALQKANNRLLKILLEVVKTTAAVEETIGRHVLGILDRSSKSQSSASLIWRSEAEASVKSCVHEEHTRVTDESIPSYSGSDMPRNDINMWSKVTEEGTELSQRLVRSGFAGTEIDPENEELMLNISSRLQAAVEKLLEAISETSSQLEHAKVTQTELMRESFRQKQEATESLKCQEELRERLHEESRAREQLAVELSKAEGVIDGYADEKTLFERQIQEKTDIIDRLEQELLCASNRLQELEAEQQQIQEERELLSRQKEAMKAEAGPVEQQLLQETEKLMKEKLEVQCQAEKVRDDLQKQVKALEIDVEEQVSRFIELEQEKNTELMDLRQQNQALEKQLEKMRKFLDEQAIDREHERDVFQQEIQKLEQQLKVVPRFQPISEHQTREVEQLANHLKEKTDKCSELLLSKEQLQRDIQERNEEIEKLEFRVRELEQALLVSADTFQKVEDRKHFGAVEAKPELSLEVQLQAERDAIDRKEKEITNLEEQLEQFREELENKNEEVQQLHMQLEIQKKESTTRLQELEQENKLFKDDMEKLGLAIKESDAMSTQDQHVLFGKFAQIIQEKEVEIDQLNEQVTKLQQQLKITTDNKVIEEKNELIRDLETQIECLMSDQECVKRNREEEIEQLNEVIEKLQQELANIGQKTSMNAHSLSEEADSLKHQLDVVIAEKLALEQQVETANEEMTFMKNVLKETNFKMNQLTQELFSLKRERESVEKIQSIPENSVNVAIDHLSKDKPELEVVLTEDALKSLENQTYFKSFEENGKGSIINLETRLLQLESTVSAKDLELTQCYKQIKDMQEQGQFETEMLQKKIVNLQKIVEEKVAAALVSQIQLEAVQEYAKFCQDNQTISSEPERTNIQNLNQLREDELGSDISALTLRISELESQVVEMHTSLILEKEQVEIAEKNVLEKEKKLLELQKLLEGNEKKQREKEKKRSPQDVEVLKTTTELFHSNEESGFFNELEALRAESVATKAELASYKEKAEKLQEELLVKETNMTSLQKDLSQVRDHLAEAKEKLSILEKEDETEVQESKKACMFEPLPIKLSKSIASQTDGTLKISSSNQTPQILVKNAGIQINLQSECSSEEVTEIISQFTEKIEKMQELHAAEILDMESRHISETETLKREHYVAVQLLKEECGTLKAVIQCLRSKEGSSIPELAHSDAYQTREICSSDSGSDWGQGIYLTHSQGFDIASEGRGEESESATDSFPKKIKGLLRAVHNEGMQVLSLTESPYSDGEDHSIQQVSEPWLEERKAYINTISSLKDLITKMQLQREAEVYDSSQSHESFSDWRGELLLALQQVFLEERSVLLAAFRTELTALGTTDAVGLLNCLEQRIQEQGVEYQAAMECLQKADRRSLLSEIQALHAQMNGRKITLKREQESEKPSQELLEYNIQQKQSQMLEMQVELSSMKDRATELQEQLSSEKMVVAELKSELAQTKLELETTLKAQHKHLKELEAFRLEVKDKTDEVHLLNDTLASEQKKSRELQWALEKEKAKLGRSEERDKEELEDLKFSLESQKQRNLQLNLLLEQQKQLLNESQQKIESQRMLYDAQLSEEQGRNLELQVLLESEKVRIREMSSTLDRERELHAQLQSSDGTGQSRPPLPSEDLLKELQKQLEEKHSRIVELLNETEKYKLDSLQTRQQMEKDRQVHRKTLQTEQEANTEGQKKMHELQSKVEDLQRQLEEKRQQVYKLDLEGQRLQGIMQEFQKQELEREEKRESRRILYQNLNEPTTWSLTSDRTRNWVLQQKIEGETKESNYAKLIEMNGGGTGCNHELEMIRQKLQCVASKLQVLPQKASERLQFETADDEDFIWVQENIDEIILQLQKLTGQQGEEPSLVSPSTSCGSLTERLLRQNAELTGHISQLTEEKNDLRNMVMKLEEQIRWYRQTGAGRDNSSRFSLNGGANIEAIIASEKEVWNREKLTLQKSLKRAEAEVYKLKAELRNDSLLQTLSPDSEHVTLKRIYGKYLRAESFRKALIYQKKYLLLLLGGFQECEDATLALLARMGGQPAFTDLEVITNRPKGFTRFRSAVRVSIAISRMKFLVRRWHRVTGSVSININRDGFGLNQGAEKTDSFYHSSGGLELYGEPRHTTYRSRSDLDYIRSPLPFQNRYPGTPADFNPGSLACSQLQNYDPDRALTDYITRLEALQRRLGTIQSGSTTQFHAGMRR
Predictions
Score>80(red) has high confidence and is suggested to be used for WB detection. *The prediction model is mainly based on the alignment of immunogen sequences, the results are for reference only, not as the basis of quality assurance.
High(score>80) Medium(80>score>50) Low(score<50) No confidence
Research Backgrounds
Scaffolding protein that assembles several protein kinases and phosphatases on the centrosome and Golgi apparatus. Required to maintain the integrity of the Golgi apparatus. Required for microtubule nucleation at the cis-side of the Golgi apparatus. Required for association of the centrosomes with the poles of the bipolar mitotic spindle during metaphase. In complex with PDE4DIP isoform 13/MMG8/SMYLE, recruits CAMSAP2 to the Golgi apparatus and tethers non-centrosomal minus-end microtubules to the Golgi, an important step for polarized cell movement. In complex with PDE4DIP isoform 13/MMG8/SMYLE, EB1/MAPRE1 and CDK5RAP2, contributes to microtubules nucleation and extension also from the centrosome to the cell periphery.
Associated with the N-methyl-D-aspartate receptor and is specifically found in the neuromuscular junction (NMJ) as well as in neuronal synapses, suggesting a role in the organization of postsynaptic specializations.
Golgi apparatus. Cytoplasm. Cytoplasm>Cytoskeleton>Microtubule organizing center>Centrosome.
Note: Cytoplasmic in parietal cells (PubMed:9915845). Recruited to the Golgi apparatus by GM130/GOLGA2 (PubMed:25657325). Localization at the centrosome versus Golgi apparatus may be cell line-dependent. In SKBr3 and HEK293F cells, exclusively located at the centrosome (PubMed:29162697). In HeLa, MDA-MB231 and RPE-1 cells, detected at the Golgi apparatus (PubMed:25217626, PubMed:29162697). In SK-BR-3 cells, recruited to the centrosome in the presence of CDK5RAP2 (PubMed:29162697).
Widely expressed. Isoform 4: Highly expressed in skeletal muscle and in pancreas.
RII-binding site, predicted to form an amphipathic helix, could participate in protein-protein interactions with a complementary surface on the R-subunit dimer.
Restrictive clause
Affinity Biosciences tests all products strictly. Citations are provided as a resource for additional applications that have not been validated by Affinity Biosciences. Please choose the appropriate format for each application and consult Materials and Methods sections for additional details about the use of any product in these publications.
For Research Use Only.
Not for use in diagnostic or therapeutic procedures. Not for resale. Not for distribution without written consent. Affinity Biosciences will not be held responsible for patent infringement or other violations that may occur with the use of our products. Affinity Biosciences, Affinity Biosciences Logo and all other trademarks are the property of Affinity Biosciences LTD.