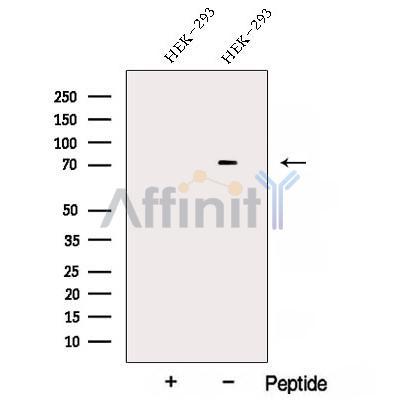
AKAP8L Antibody - #DF12818
| Product: | AKAP8L Antibody |
| Catalog: | DF12818 |
| Description: | Rabbit polyclonal antibody to AKAP8L |
| Application: | WB |
| Reactivity: | Human |
| Prediction: | Pig, Bovine, Horse, Sheep, Rabbit, Dog |
| Mol.Wt.: | 72 kDa; 72kD(Calculated). |
| Uniprot: | Q9ULX6 |
| RRID: | AB_2845779 |
Related Downloads
Protocols
Product Info
*The optimal dilutions should be determined by the end user. For optimal experimental results, antibody reuse is not recommended.
*Tips:
WB: For western blot detection of denatured protein samples. IHC: For immunohistochemical detection of paraffin sections (IHC-p) or frozen sections (IHC-f) of tissue samples. IF/ICC: For immunofluorescence detection of cell samples. ELISA(peptide): For ELISA detection of antigenic peptide.
Cite Format: Affinity Biosciences Cat# DF12818, RRID:AB_2845779.
Fold/Unfold
A kinase (PRKA) anchor protein 8 like; A kinase anchor protein 8 like; AKAP 8L; AKAP8 like protein; DKFZp434L0650; HA95; HAP95; Helicase A binding protein 95; Homologous to AKAP95 protein; NAKAP; NAKAP95; Neighbor of A kinase anchoring protein 95; Neighbor of AKAP95;
Immunogens
A synthesized peptide derived from human AKAP8L, corresponding to a region within N-terminal amino acids.
- Q9ULX6 AKP8L_HUMAN:
- Protein BLAST With
- NCBI/
- ExPASy/
- Uniprot
MSYTGFVQGSETTLQSTYSDTSAQPTCDYGYGTWNSGTNRGYEGYGYGYGYGQDNTTNYGYGMATSHSWEMPSSDTNANTSASGSASADSVLSRINQRLDMVPHLETDMMQGGVYGSGGERYDSYESCDSRAVLSERDLYRSGYDYSELDPEMEMAYEGQYDAYRDQFRMRGNDTFGPRAQGWARDARSGRPMASGYGRMWEDPMGARGQCMSGASRLPSLFSQNIIPEYGMFQGMRGGGAFPGGSRFGFGFGNGMKQMRRTWKTWTTADFRTKKKKRKQGGSPDEPDSKATRTDCSDNSDSDNDEGTEGEATEGLEGTEAVEKGSRVDGEDEEGKEDGREEGKEDPEKGALTTQDENGQTKRKLQAGKKSQDKQKKRQRDRMVERIQFVCSLCKYRTFYEDEMASHLDSKFHKEHFKYVGTKLPKQTADFLQEYVTNKTKKTEELRKTVEDLDGLIQQIYRDQDLTQEIAMEHFVKKVEAAHCAACDLFIPMQFGIIQKHLKTMDHNRNRRLMMEQSKKSSLMVARSILNNKLISKKLERYLKGENPFTDSPEEEKEQEEAEGGALDEGAQGEAAGISEGAEGVPAQPPVPPEPAPGAVSPPPPPPPEEEEEGAVPLLGGALQRQIRGIPGLDVEDDEEGGGGAP
Predictions
Score>80(red) has high confidence and is suggested to be used for WB detection. *The prediction model is mainly based on the alignment of immunogen sequences, the results are for reference only, not as the basis of quality assurance.
High(score>80) Medium(80>score>50) Low(score<50) No confidence
Research Backgrounds
Could play a role in constitutive transport element (CTE)-mediated gene expression by association with DHX9. Increases CTE-dependent nuclear unspliced mRNA export. Proposed to target PRKACA to the nucleus but does not seem to be implicated in the binding of regulatory subunit II of PKA. May be involved in nuclear envelope breakdown and chromatin condensation. May be involved in anchoring nuclear membranes to chromatin in interphase and in releasing membranes from chromating at mitosis. May regulate the initiation phase of DNA replication when associated with TMPO isoform Beta. Required for cell cycle G2/M transition and histone deacetylation during mitosis. In mitotic cells recruits HDAC3 to the vicinity of chromatin leading to deacetylation and subsequent phosphorylation at 'Ser-10' of histone H3; in this function seems to act redundantly with AKAP8. May be involved in regulation of pre-mRNA splicing.
(Microbial infection) In case of EBV infection, may target PRKACA to EBNA-LP-containing nuclear sites to modulate transcription from specific promoters.
(Microbial infection) Can synergize with DHX9 to activate the CTE-mediated gene expression of type D retroviruses.
(Microbial infection) In case of HIV-1 infection, involved in the DHX9-promoted annealing of host tRNA(Lys3) to viral genomic RNA as a primer in reverse transcription; in vitro negatively regulates DHX9 annealing activity.
Phosphorylated on serine or threonine residues possibly by PKA; probably modulating the interaction with TMPO isoform Beta.
Nucleus. Nucleus matrix. Nucleus speckle. Nucleus>PML body. Cytoplasm.
Note: Colocalizes with PRPF40A in the nuclear matrix (PubMed:16391387). Nuclear at steady state but shuttles between the nucleus and cytoplasm (PubMed:10748171). The shuttling property has been questioned (PubMed:11034899). Colocalizes with EBNA-LP in PML bodies (PubMed:11884601).
Ubiquitously expressed. Expressed in the brain cortex (at protein level).
Belongs to the AKAP95 family.
Research Fields
· Human Diseases > Infectious diseases: Viral > Epstein-Barr virus infection.
Restrictive clause
Affinity Biosciences tests all products strictly. Citations are provided as a resource for additional applications that have not been validated by Affinity Biosciences. Please choose the appropriate format for each application and consult Materials and Methods sections for additional details about the use of any product in these publications.
For Research Use Only.
Not for use in diagnostic or therapeutic procedures. Not for resale. Not for distribution without written consent. Affinity Biosciences will not be held responsible for patent infringement or other violations that may occur with the use of our products. Affinity Biosciences, Affinity Biosciences Logo and all other trademarks are the property of Affinity Biosciences LTD.