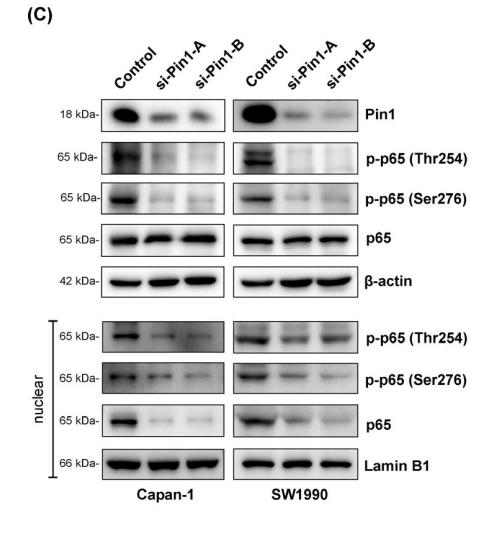
AFfirm™ Vimentin mouse monoclonal Antibody - #BF8006
| Product: | Vimentin mouse monoclonal Antibody |
| Catalog: | BF8006 |
| Description: | Mouse monoclonal antibody to Vimentin |
| Application: | WB IHC IF/ICC |
| Cited expt.: | WB, IHC, IF/ICC |
| Reactivity: | Human, Mouse, Rat |
| Mol.Wt.: | 53kDa; 54kD(Calculated). |
| Uniprot: | P08670 |
| RRID: | AB_2847777 |
Product Info
*The optimal dilutions should be determined by the end user. For optimal experimental results, antibody reuse is not recommended.
*Tips:
WB: For western blot detection of denatured protein samples. IHC: For immunohistochemical detection of paraffin sections (IHC-p) or frozen sections (IHC-f) of tissue samples. IF/ICC: For immunofluorescence detection of cell samples. ELISA(peptide): For ELISA detection of antigenic peptide.
Cite Format: Affinity Biosciences Cat# BF8006, RRID:AB_2847777.
Fold/Unfold
CTRCT30; Epididymis luminal protein 113; FLJ36605; HEL113; VIM; VIME_HUMAN; Vimentin;
Immunogens
Purified recombinant fragment of human Vimentin expressed in E. Coli.
Highly expressed in fibroblasts, some expression in T- and B-lymphocytes, and little or no expression in Burkitt's lymphoma cell lines. Expressed in many hormone-independent mammary carcinoma cell lines.
- P08670 VIME_HUMAN:
- Protein BLAST With
- NCBI/
- ExPASy/
- Uniprot
MSTRSVSSSSYRRMFGGPGTASRPSSSRSYVTTSTRTYSLGSALRPSTSRSLYASSPGGVYATRSSAVRLRSSVPGVRLLQDSVDFSLADAINTEFKNTRTNEKVELQELNDRFANYIDKVRFLEQQNKILLAELEQLKGQGKSRLGDLYEEEMRELRRQVDQLTNDKARVEVERDNLAEDIMRLREKLQEEMLQREEAENTLQSFRQDVDNASLARLDLERKVESLQEEIAFLKKLHEEEIQELQAQIQEQHVQIDVDVSKPDLTAALRDVRQQYESVAAKNLQEAEEWYKSKFADLSEAANRNNDALRQAKQESTEYRRQVQSLTCEVDALKGTNESLERQMREMEENFAVEAANYQDTIGRLQDEIQNMKEEMARHLREYQDLLNVKMALDIEIATYRKLLEGEESRISLPLPNFSSLNLRETNLDSLPLVDTHSKRTLLIKTVETRDGQVINETSQHHDDLE
Research Backgrounds
Vimentins are class-III intermediate filaments found in various non-epithelial cells, especially mesenchymal cells. Vimentin is attached to the nucleus, endoplasmic reticulum, and mitochondria, either laterally or terminally.
Involved with LARP6 in the stabilization of type I collagen mRNAs for CO1A1 and CO1A2.
Filament disassembly during mitosis is promoted by phosphorylation at Ser-55 as well as by nestin (By similarity). One of the most prominent phosphoproteins in various cells of mesenchymal origin. Phosphorylation is enhanced during cell division, at which time vimentin filaments are significantly reorganized. Phosphorylation by PKN1 inhibits the formation of filaments. Phosphorylated at Ser-56 by CDK5 during neutrophil secretion in the cytoplasm. Phosphorylated by STK33. Phosphorylated on tyrosine residues by SRMS.
O-glycosylated during cytokinesis at sites identical or close to phosphorylation sites, this interferes with the phosphorylation status.
S-nitrosylation is induced by interferon-gamma and oxidatively-modified low-densitity lipoprotein (LDL(ox)) possibly implicating the iNOS-S100A8/9 transnitrosylase complex.
Cytoplasm. Cytoplasm>Cytoskeleton. Nucleus matrix.
Highly expressed in fibroblasts, some expression in T- and B-lymphocytes, and little or no expression in Burkitt's lymphoma cell lines. Expressed in many hormone-independent mammary carcinoma cell lines.
The central alpha-helical coiled-coil IF rod domain mediates elementary homodimerization.
The [IL]-x-C-x-x-[DE] motif is a proposed target motif for cysteine S-nitrosylation mediated by the iNOS-S100A8/A9 transnitrosylase complex.
Belongs to the intermediate filament family.
Research Fields
· Human Diseases > Infectious diseases: Viral > Epstein-Barr virus infection.
· Human Diseases > Cancers: Overview > MicroRNAs in cancer.
References
Application: IHC Species: mouse Sample: tumor
Application: IF/ICC Species: human Sample: HCC cells
Application: IHC Species: Rat Sample: lung tissue
Application: WB Species: Mouse Sample: MLE-12 cells
Application: WB Species: Mouse Sample: GC cells
Restrictive clause
Affinity Biosciences tests all products strictly. Citations are provided as a resource for additional applications that have not been validated by Affinity Biosciences. Please choose the appropriate format for each application and consult Materials and Methods sections for additional details about the use of any product in these publications.
For Research Use Only.
Not for use in diagnostic or therapeutic procedures. Not for resale. Not for distribution without written consent. Affinity Biosciences will not be held responsible for patent infringement or other violations that may occur with the use of our products. Affinity Biosciences, Affinity Biosciences Logo and all other trademarks are the property of Affinity Biosciences LTD.