VE-Cadherin Antibody - #AF6265
| Product: | VE-Cadherin Antibody |
| Catalog: | AF6265 |
| Description: | Rabbit polyclonal antibody to VE-Cadherin |
| Application: | WB IHC IF/ICC |
| Cited expt.: | WB, IHC, IF/ICC |
| Reactivity: | Human, Mouse |
| Prediction: | Pig, Zebrafish, Bovine, Horse, Sheep, Dog, Chicken |
| Mol.Wt.: | 120kDa; 88kD(Calculated). |
| Uniprot: | P33151 |
| RRID: | AB_2835123 |
Product Info
*The optimal dilutions should be determined by the end user. For optimal experimental results, antibody reuse is not recommended.
*Tips:
WB: For western blot detection of denatured protein samples. IHC: For immunohistochemical detection of paraffin sections (IHC-p) or frozen sections (IHC-f) of tissue samples. IF/ICC: For immunofluorescence detection of cell samples. ELISA(peptide): For ELISA detection of antigenic peptide.
Cite Format: Affinity Biosciences Cat# AF6265, RRID:AB_2835123.
Fold/Unfold
7B 4; 7B4; 7B4 antigen; CADH5_HUMAN; Cadherin 5; Cadherin 5 type 2; Cadherin 5, type 2 (vascular endothelium); Cadherin 5, type 2, VE cadherin (vascular epithelium); cadherin, vascular endothelial; cadherin, vascular endothelial, 1; Cadherin-5; Cadherin5; CD 144; CD144; CD144 antigen; CDH 5; CDH5; CDH5 protein; Endothelial specific cadherin; FLJ17376; OTTHUMP00000174777; Vascular endothelial cadherin; Vascular epithelium cadherin; VE Cad; VE-cadherin; VEC;
Immunogens
A synthesized peptide derived from human VE-Cadherin, corresponding to a region within C-terminal amino acids.
- P33151 CADH5_HUMAN:
- Protein BLAST With
- NCBI/
- ExPASy/
- Uniprot
MQRLMMLLATSGACLGLLAVAAVAAAGANPAQRDTHSLLPTHRRQKRDWIWNQMHIDEEKNTSLPHHVGKIKSSVSRKNAKYLLKGEYVGKVFRVDAETGDVFAIERLDRENISEYHLTAVIVDKDTGENLETPSSFTIKVHDVNDNWPVFTHRLFNASVPESSAVGTSVISVTAVDADDPTVGDHASVMYQILKGKEYFAIDNSGRIITITKSLDREKQARYEIVVEARDAQGLRGDSGTATVLVTLQDINDNFPFFTQTKYTFVVPEDTRVGTSVGSLFVEDPDEPQNRMTKYSILRGDYQDAFTIETNPAHNEGIIKPMKPLDYEYIQQYSFIVEATDPTIDLRYMSPPAGNRAQVIINITDVDEPPIFQQPFYHFQLKENQKKPLIGTVLAMDPDAARHSIGYSIRRTSDKGQFFRVTKKGDIYNEKELDREVYPWYNLTVEAKELDSTGTPTGKESIVQVHIEVLDENDNAPEFAKPYQPKVCENAVHGQLVLQISAIDKDITPRNVKFKFILNTENNFTLTDNHDNTANITVKYGQFDREHTKVHFLPVVISDNGMPSRTGTSTLTVAVCKCNEQGEFTFCEDMAAQVGVSIQAVVAILLCILTITVITLLIFLRRRLRKQARAHGKSVPEIHEQLVTYDEEGGGEMDTTSYDVSVLNSVRRGGAKPPRPALDARPSLYAQVQKPPRHAPGAHGGPGEMAAMIEVKKDEADHDGDGPPYDTLHIYGYEGSESIAESLSSLGTDSSDSDVDYDFLNDWGPRFKMLAELYGSDPREELLY
Predictions
Score>80(red) has high confidence and is suggested to be used for WB detection. *The prediction model is mainly based on the alignment of immunogen sequences, the results are for reference only, not as the basis of quality assurance.
High(score>80) Medium(80>score>50) Low(score<50) No confidence
Research Backgrounds
Cadherins are calcium-dependent cell adhesion proteins (By similarity). They preferentially interact with themselves in a homophilic manner in connecting cells; cadherins may thus contribute to the sorting of heterogeneous cell types. This cadherin may play a important role in endothelial cell biology through control of the cohesion and organization of the intercellular junctions (By similarity). It associates with alpha-catenin forming a link to the cytoskeleton. Acts in concert with KRIT1 and MPP5 to establish and maintain correct endothelial cell polarity and vascular lumen (By similarity). These effects are mediated by recruitment and activation of the Par polarity complex and RAP1B. Required for activation of PRKCZ and for the localization of phosphorylated PRKCZ, PARD3, TIAM1 and RAP1B to the cell junction.
Phosphorylated on tyrosine residues by KDR/VEGFR-2. Dephosphorylated by PTPRB (By similarity).
O-glycosylated.
Cell junction. Cell membrane>Single-pass type I membrane protein.
Note: Found at cell-cell boundaries and probably at cell-matrix boundaries. KRIT1 and CDH5 reciprocally regulate their localization to endothelial cell-cell junctions.
Endothelial tissues and brain.
Three calcium ions are usually bound at the interface of each cadherin domain and rigidify the connections, imparting a strong curvature to the full-length ectodomain.
Research Fields
· Environmental Information Processing > Signaling molecules and interaction > Cell adhesion molecules (CAMs). (View pathway)
· Organismal Systems > Immune system > Leukocyte transendothelial migration. (View pathway)
References
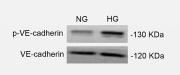
Application: WB Species: human Sample: U87 and U251 cells
Application: WB Species: human Sample: PLC-PRF-5 cells
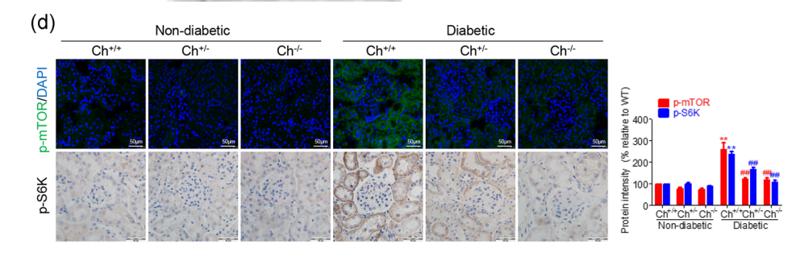
Application: IHC Species: human Sample: HCC
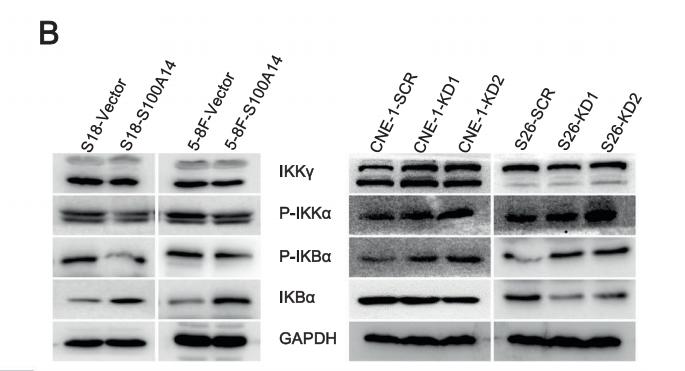
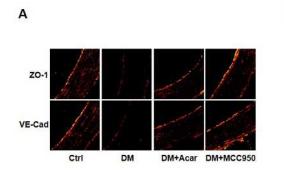
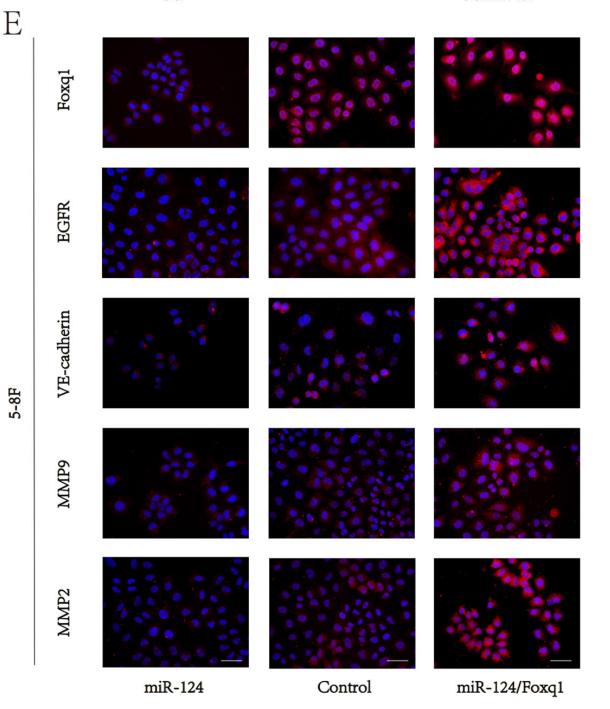
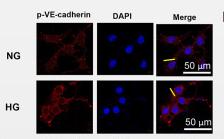
Application: IF/ICC Species: mouse Sample: 5–8F cells
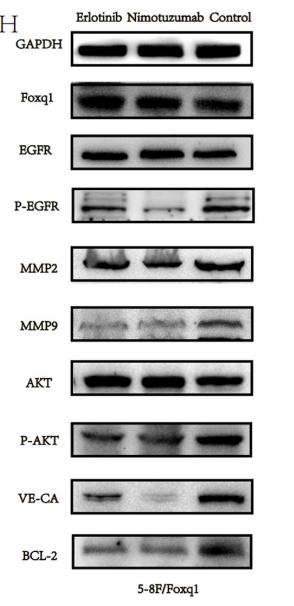
Application: WB Species: mouse Sample: 5–8F cells
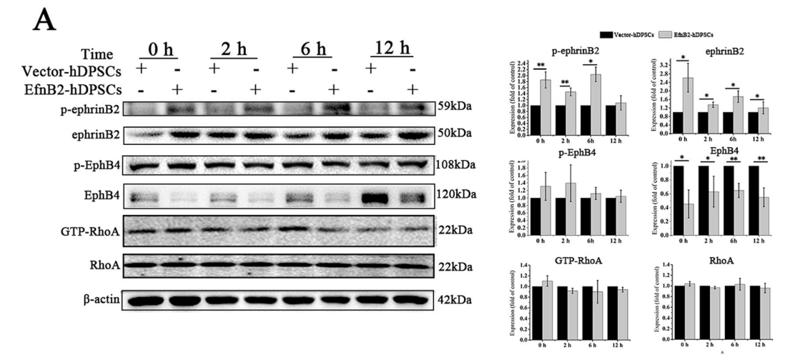
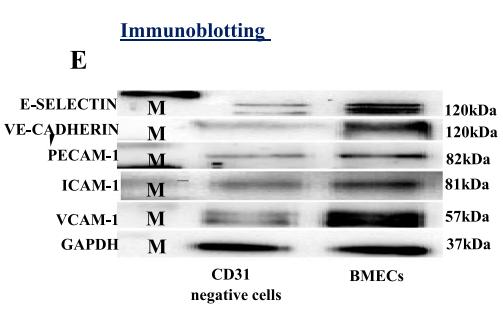
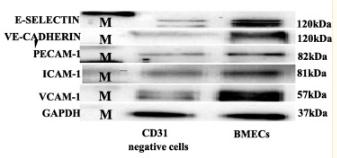
Application: WB Species: mouse Sample: primary bone marrow endothelial cells
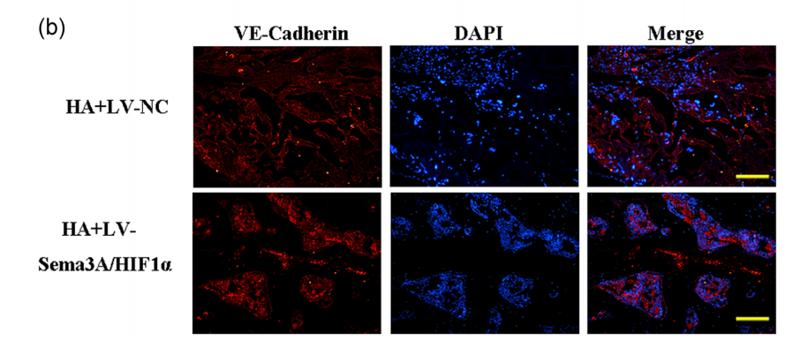
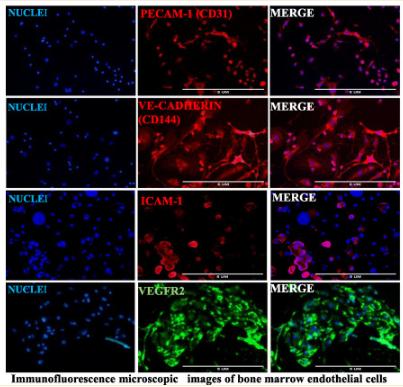
Application: IF/ICC Species: mouse Sample: primary bone marrow endothelial cells
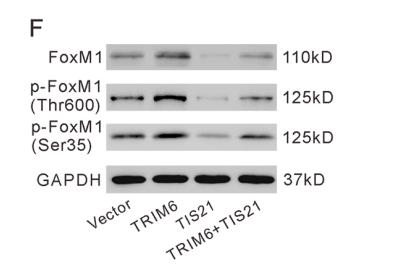
Application: WB Species: Mice Sample: bMECs
Application: IF/ICC Species: Mice Sample: BMECs
Application: IHC Species: Mouse Sample:
Application: WB Species: rat Sample: endothelial cells
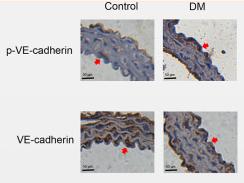
Application: IF/ICC Species: rat Sample: arterial endothelium
Restrictive clause
Affinity Biosciences tests all products strictly. Citations are provided as a resource for additional applications that have not been validated by Affinity Biosciences. Please choose the appropriate format for each application and consult Materials and Methods sections for additional details about the use of any product in these publications.
For Research Use Only.
Not for use in diagnostic or therapeutic procedures. Not for resale. Not for distribution without written consent. Affinity Biosciences will not be held responsible for patent infringement or other violations that may occur with the use of our products. Affinity Biosciences, Affinity Biosciences Logo and all other trademarks are the property of Affinity Biosciences LTD.