14-3-3 zeta/delta Antibody - #AF0097
| Product: | 14-3-3 zeta/delta Antibody |
| Catalog: | AF0097 |
| Description: | Rabbit polyclonal antibody to 14-3-3 zeta/delta |
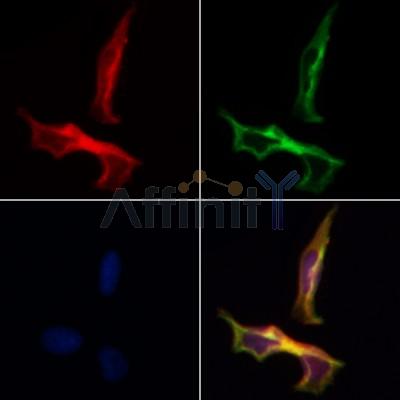
| Application: | WB IF/ICC |
| Reactivity: | Human, Mouse, Rat |
| Prediction: | Pig, Zebrafish, Bovine, Horse, Sheep, Rabbit, Dog, Chicken |
| Mol.Wt.: | 28kDa; 28kD(Calculated). |
| Uniprot: | P63104 | P31946 |
| RRID: | AB_2833278 |
Related Downloads
Protocols
Product Info
*The optimal dilutions should be determined by the end user. For optimal experimental results, antibody reuse is not recommended.
*Tips:
WB: For western blot detection of denatured protein samples. IHC: For immunohistochemical detection of paraffin sections (IHC-p) or frozen sections (IHC-f) of tissue samples. IF/ICC: For immunofluorescence detection of cell samples. ELISA(peptide): For ELISA detection of antigenic peptide.
Cite Format: Affinity Biosciences Cat# AF0097, RRID:AB_2833278.
Fold/Unfold
14 3 3 delta; 14 3 3 protein zeta/delta; 14 3 3 protein/cytosolic phospholipase A2; 14 3 3 zeta; 14-3-3 protein zeta/delta; 1433Z_HUMAN; Epididymis luminal protein 4; Epididymis secretory protein Li 3; HEL S 3; HEL4; KCIP-1; KCIP1; MGC111427; MGC126532; MGC138156; Phospholipase A2; Protein kinase C inhibitor protein 1; Tyrosine 3 monooxygenase/tryptophan 5 monooxygenase activation protein, delta polypeptide; Tyrosine 3 monooxygenase/tryptophan 5 monooxygenase activation protein, zeta; Tyrosine 3 monooxygenase/tryptophan 5 monooxygenase activation protein, zeta polypeptide; Tyrosine 3/tryptophan 5 monooxygenase activation protein, zeta polypeptide; YWHAD; YWHAZ; 14 3 3 alpha; 14 3 3 protein beta/alpha; 14-3-3 protein beta/alpha; 1433B_HUMAN; Brain protein 14 3 3 beta isoform; GW128; HS 1; KCIP-1; KCIP1; N-terminally processed; Protein 1054; Protein kinase C inhibitor protein 1; YWHAA; YWHAB;
Immunogens
A synthesized peptide derived from human 14-3-3 zeta/delta, corresponding to a region within the internal amino acids.
- P63104 1433Z_HUMAN:
- Protein BLAST With
- NCBI/
- ExPASy/
- Uniprot
MDKNELVQKAKLAEQAERYDDMAACMKSVTEQGAELSNEERNLLSVAYKNVVGARRSSWRVVSSIEQKTEGAEKKQQMAREYREKIETELRDICNDVLSLLEKFLIPNASQAESKVFYLKMKGDYYRYLAEVAAGDDKKGIVDQSQQAYQEAFEISKKEMQPTHPIRLGLALNFSVFYYEILNSPEKACSLAKTAFDEAIAELDTLSEESYKDSTLIMQLLRDNLTLWTSDTQGDEAEAGEGGEN
- P31946 1433B_HUMAN:
- Protein BLAST With
- NCBI/
- ExPASy/
- Uniprot
MTMDKSELVQKAKLAEQAERYDDMAAAMKAVTEQGHELSNEERNLLSVAYKNVVGARRSSWRVISSIEQKTERNEKKQQMGKEYREKIEAELQDICNDVLELLDKYLIPNATQPESKVFYLKMKGDYFRYLSEVASGDNKQTTVSNSQQAYQEAFEISKKEMQPTHPIRLGLALNFSVFYYEILNSPEKACSLAKTAFDEAIAELDTLNEESYKDSTLIMQLLRDNLTLWTSENQGDEGDAGEGEN
Predictions
Score>80(red) has high confidence and is suggested to be used for WB detection. *The prediction model is mainly based on the alignment of immunogen sequences, the results are for reference only, not as the basis of quality assurance.
High(score>80) Medium(80>score>50) Low(score<50) No confidence
Research Backgrounds
Adapter protein implicated in the regulation of a large spectrum of both general and specialized signaling pathways. Binds to a large number of partners, usually by recognition of a phosphoserine or phosphothreonine motif. Binding generally results in the modulation of the activity of the binding partner. Induces ARHGEF7 activity on RAC1 as well as lamellipodia and membrane ruffle formation. In neurons, regulates spine maturation through the modulation of ARHGEF7 activity (By similarity).
The delta, brain-specific form differs from the zeta form in being phosphorylated (By similarity). Phosphorylation on Ser-184 by MAPK8; promotes dissociation of BAX and translocation of BAX to mitochondria. Phosphorylation on Thr-232; inhibits binding of RAF1. Phosphorylated on Ser-58 by PKA and protein kinase C delta type catalytic subunit in a sphingosine-dependent fashion. Phosphorylation on Ser-58 by PKA; disrupts homodimerization and heterodimerization with YHAE and TP53.
Cytoplasm. Melanosome.
Note: Located to stage I to stage IV melanosomes.
Belongs to the 14-3-3 family.
Adapter protein implicated in the regulation of a large spectrum of both general and specialized signaling pathways. Binds to a large number of partners, usually by recognition of a phosphoserine or phosphothreonine motif. Binding generally results in the modulation of the activity of the binding partner. Negative regulator of osteogenesis. Blocks the nuclear translocation of the phosphorylated form (by AKT1) of SRPK2 and antagonizes its stimulatory effect on cyclin D1 expression resulting in blockage of neuronal apoptosis elicited by SRPK2. Negative regulator of signaling cascades that mediate activation of MAP kinases via AKAP13.
The alpha, brain-specific form differs from the beta form in being phosphorylated. Phosphorylated on Ser-60 by protein kinase C delta type catalytic subunit in a sphingosine-dependent fashion.
Cytoplasm. Melanosome.
Note: Identified by mass spectrometry in melanosome fractions from stage I to stage IV.
Vacuole membrane.
Note: (Microbial infection) Upon infection with Chlamydia trachomatis, this protein is associated with the pathogen-containing vacuole membrane where it colocalizes with IncG.
Belongs to the 14-3-3 family.
Research Fields
· Cellular Processes > Cell growth and death > Cell cycle. (View pathway)
· Cellular Processes > Cell growth and death > Oocyte meiosis. (View pathway)
· Environmental Information Processing > Signal transduction > PI3K-Akt signaling pathway. (View pathway)
· Environmental Information Processing > Signal transduction > Hippo signaling pathway. (View pathway)
· Human Diseases > Infectious diseases: Bacterial > Pathogenic Escherichia coli infection.
· Human Diseases > Infectious diseases: Viral > Hepatitis B.
· Human Diseases > Infectious diseases: Viral > Epstein-Barr virus infection.
· Human Diseases > Cancers: Overview > Viral carcinogenesis.
Restrictive clause
Affinity Biosciences tests all products strictly. Citations are provided as a resource for additional applications that have not been validated by Affinity Biosciences. Please choose the appropriate format for each application and consult Materials and Methods sections for additional details about the use of any product in these publications.
For Research Use Only.
Not for use in diagnostic or therapeutic procedures. Not for resale. Not for distribution without written consent. Affinity Biosciences will not be held responsible for patent infringement or other violations that may occur with the use of our products. Affinity Biosciences, Affinity Biosciences Logo and all other trademarks are the property of Affinity Biosciences LTD.