CD9 Antibody - #AF5139
| Product: | CD9 Antibody |
| Catalog: | AF5139 |
| Description: | Rabbit polyclonal antibody to CD9 |
| Application: | WB IHC IF/ICC |
| Cited expt.: | WB |
| Reactivity: | Human, Mouse, Rat |
| Prediction: | Pig, Bovine, Horse, Sheep, Rabbit, Dog |
| Mol.Wt.: | 20~35kD; 25kD(Calculated). |
| Uniprot: | P21926 |
| RRID: | AB_2837625 |
Related Downloads
Protocols
Product Info
*The optimal dilutions should be determined by the end user. For optimal experimental results, antibody reuse is not recommended.
*Tips:
WB: For western blot detection of denatured protein samples. IHC: For immunohistochemical detection of paraffin sections (IHC-p) or frozen sections (IHC-f) of tissue samples. IF/ICC: For immunofluorescence detection of cell samples. ELISA(peptide): For ELISA detection of antigenic peptide.
Cite Format: Affinity Biosciences Cat# AF5139, RRID:AB_2837625.
Fold/Unfold
Tetraspanin 29; 5H9; 5H9 antigen; Antigen defined by monoclonal antibody 602 29; Antigen defined by monoclonal antibody 60229; BA-2/p24 antigen; BA2; BTCC 1; BTCC1; CD9; CD9 antigen; CD9 antigen p24; CD9 molecule; CD9_HUMAN; Cell growth inhibiting gene 2 protein; Cell growth-inhibiting gene 2 protein; DRAP 27; DRAP27; GIG2; Growth inhibiting gene 2 protein; Leukocyte antigen MIC3; MIC3; Motility related protein; Motility-related protein; MRP 1; MRP-1; MRP1; p24; p24 antigen; Tetraspanin-29; Tspan 29; Tspan-29; TSPAN29;
Immunogens
A synthesized peptide derived from human CD9, corresponding to a region within the internal amino acids.
Detected in platelets (at protein level) (PubMed:19640571). Expressed by a variety of hematopoietic and epithelial cells (PubMed:19640571).
- P21926 CD9_HUMAN:
- Protein BLAST With
- NCBI/
- ExPASy/
- Uniprot
MPVKGGTKCIKYLLFGFNFIFWLAGIAVLAIGLWLRFDSQTKSIFEQETNNNNSSFYTGVYILIGAGALMMLVGFLGCCGAVQESQCMLGLFFGFLLVIFAIEIAAAIWGYSHKDEVIKEVQEFYKDTYNKLKTKDEPQRETLKAIHYALNCCGLAGGVEQFISDICPKKDVLETFTVKSCPDAIKEVFDNKFHIIGAVGIGIAVVMIFGMIFSMILCCAIRRNREMV
Predictions
Score>80(red) has high confidence and is suggested to be used for WB detection. *The prediction model is mainly based on the alignment of immunogen sequences, the results are for reference only, not as the basis of quality assurance.
High(score>80) Medium(80>score>50) Low(score<50) No confidence
Research Backgrounds
Integral membrane protein associated with integrins, which regulates different processes, such as sperm-egg fusion, platelet activation and aggregation, and cell adhesion. Present at the cell surface of oocytes and plays a key role in sperm-egg fusion, possibly by organizing multiprotein complexes and the morphology of the membrane required for the fusion (By similarity). In myoblasts, associates with CD81 and PTGFRN and inhibits myotube fusion during muscle regeneration (By similarity). In macrophages, associates with CD81 and beta-1 and beta-2 integrins, and prevents macrophage fusion into multinucleated giant cells specialized in ingesting complement-opsonized large particles. Also prevents the fusion between mononuclear cell progenitors into osteoclasts in charge of bone resorption (By similarity). Acts as a receptor for PSG17 (By similarity). Involved in platelet activation and aggregation. Regulates paranodal junction formation (By similarity). Involved in cell adhesion, cell motility and tumor metastasis.
Palmitoylated at a low, basal level in unstimulated platelets. The level of palmitoylation increases when platelets are activated by thrombin (in vitro). The protein exists in three forms with molecular masses between 22 and 27 kDa, and is known to carry covalently linked fatty acids.
Cell membrane>Multi-pass membrane protein. Membrane>Multi-pass membrane protein. Secreted>Extracellular exosome.
Note: Present at the cell surface of oocytes. Accumulates in the adhesion area between the sperm and egg following interaction between IZUMO1 and its receptor IZUMO1R/JUNO.
Detected in platelets (at protein level). Expressed by a variety of hematopoietic and epithelial cells.
Belongs to the tetraspanin (TM4SF) family.
Research Fields
· Organismal Systems > Immune system > Hematopoietic cell lineage. (View pathway)
References
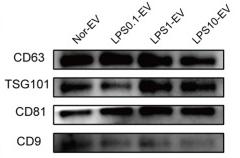
Application: WB Species: Mouse Sample:
Application: WB Species: Human Sample: HEK293T cells
Application: WB Species: Human Sample: HEK293T cells
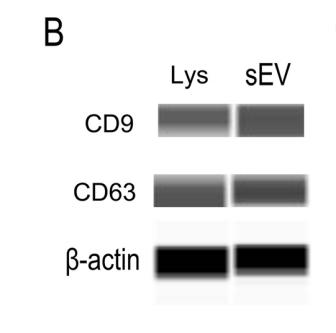
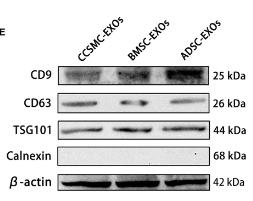
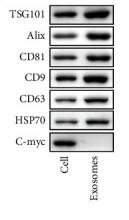
Application: WB Species: Human Sample: DPSCs-sEV
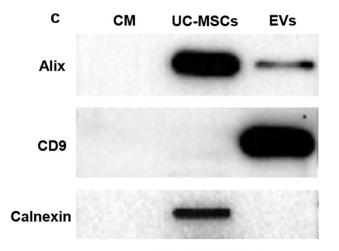
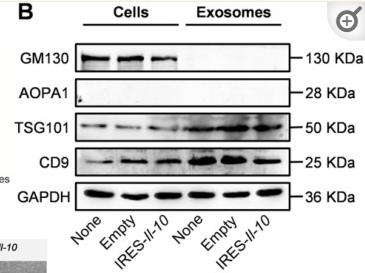
Application: WB Species: Human Sample: SCAPs
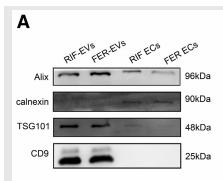
Application: WB Species: Human Sample: RIF-EVs and FER-EVs.
Restrictive clause
Affinity Biosciences tests all products strictly. Citations are provided as a resource for additional applications that have not been validated by Affinity Biosciences. Please choose the appropriate format for each application and consult Materials and Methods sections for additional details about the use of any product in these publications.
For Research Use Only.
Not for use in diagnostic or therapeutic procedures. Not for resale. Not for distribution without written consent. Affinity Biosciences will not be held responsible for patent infringement or other violations that may occur with the use of our products. Affinity Biosciences, Affinity Biosciences Logo and all other trademarks are the property of Affinity Biosciences LTD.