PDK4 Antibody - #DF7169
| Product: | PDK4 Antibody |
| Catalog: | DF7169 |
| Description: | Rabbit polyclonal antibody to PDK4 |
| Application: | WB IHC IF/ICC |
| Cited expt.: | WB, IF/ICC |
| Reactivity: | Human, Mouse, Rat |
| Prediction: | Pig, Zebrafish, Bovine, Horse, Sheep, Rabbit, Dog, Chicken |
| Mol.Wt.: | 46kDa; 46kD(Calculated). |
| Uniprot: | Q16654 |
| RRID: | AB_2839121 |
Related Downloads
Protocols
Product Info
*The optimal dilutions should be determined by the end user. For optimal experimental results, antibody reuse is not recommended.
*Tips:
WB: For western blot detection of denatured protein samples. IHC: For immunohistochemical detection of paraffin sections (IHC-p) or frozen sections (IHC-f) of tissue samples. IF/ICC: For immunofluorescence detection of cell samples. ELISA(peptide): For ELISA detection of antigenic peptide.
Cite Format: Affinity Biosciences Cat# DF7169, RRID:AB_2839121.
Fold/Unfold
[Pyruvate dehydrogenase [lipoamide]] kinase isozyme 4; FLJ40832; mitochondrial; Pdk4; PDK4_HUMAN; Pyruvate dehydrogenase [lipoamide] kinase isozyme 4 mitochondrial; Pyruvate dehydrogenase kinase 4; Pyruvate dehydrogenase kinase isoenzyme 4; Pyruvate dehydrogenase kinase isoform 4; Pyruvate dehydrogenase kinase isozyme 4; Pyruvate dehydrogenase kinase isozyme 4 mitochondrial;
Immunogens
A synthesized peptide derived from human PDK4, corresponding to a region within C-terminal amino acids.
- Q16654 PDK4_HUMAN:
- Protein BLAST With
- NCBI/
- ExPASy/
- Uniprot
MKAARFVLRSAGSLNGAGLVPREVEHFSRYSPSPLSMKQLLDFGSENACERTSFAFLRQELPVRLANILKEIDILPTQLVNTSSVQLVKSWYIQSLMDLVEFHEKSPDDQKALSDFVDTLIKVRNRHHNVVPTMAQGIIEYKDACTVDPVTNQNLQYFLDRFYMNRISTRMLMNQHILIFSDSQTGNPSHIGSIDPNCDVVAVVQDAFECSRMLCDQYYLSSPELKLTQVNGKFPDQPIHIVYVPSHLHHMLFELFKNAMRATVEHQENQPSLTPIEVIVVLGKEDLTIKISDRGGGVPLRIIDRLFSYTYSTAPTPVMDNSRNAPLAGFGYGLPISRLYAKYFQGDLNLYSLSGYGTDAIIYLKALSSESIEKLPVFNKSAFKHYQMSSEADDWCIPSREPKNLAKEVAM
Predictions
Score>80(red) has high confidence and is suggested to be used for WB detection. *The prediction model is mainly based on the alignment of immunogen sequences, the results are for reference only, not as the basis of quality assurance.
High(score>80) Medium(80>score>50) Low(score<50) No confidence
Research Backgrounds
Kinase that plays a key role in regulation of glucose and fatty acid metabolism and homeostasis via phosphorylation of the pyruvate dehydrogenase subunits PDHA1 and PDHA2. This inhibits pyruvate dehydrogenase activity, and thereby regulates metabolite flux through the tricarboxylic acid cycle, down-regulates aerobic respiration and inhibits the formation of acetyl-coenzyme A from pyruvate. Inhibition of pyruvate dehydrogenase decreases glucose utilization and increases fat metabolism in response to prolonged fasting and starvation. Plays an important role in maintaining normal blood glucose levels under starvation, and is involved in the insulin signaling cascade. Via its regulation of pyruvate dehydrogenase activity, plays an important role in maintaining normal blood pH and in preventing the accumulation of ketone bodies under starvation. In the fed state, mediates cellular responses to glucose levels and to a high-fat diet. Regulates both fatty acid oxidation and de novo fatty acid biosynthesis. Plays a role in the generation of reactive oxygen species. Protects detached epithelial cells against anoikis. Plays a role in cell proliferation via its role in regulating carbohydrate and fatty acid metabolism.
Mitochondrion matrix.
Ubiquitous; highest levels of expression in heart and skeletal muscle.
Belongs to the PDK/BCKDK protein kinase family.
References
Application: WB Species: Rat Sample:
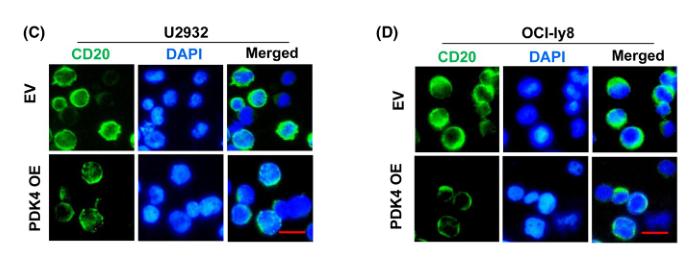
Application: IF/ICC Species: Human Sample: DLBCL cells
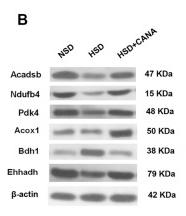
Application: WB Species: Human Sample: DLBCL cells
Application: WB Species: Human Sample: DLBCL cells
Application: IF/ICC Species: Human Sample: DLBCL cells
Application: WB Species: Mouse Sample:
Restrictive clause
Affinity Biosciences tests all products strictly. Citations are provided as a resource for additional applications that have not been validated by Affinity Biosciences. Please choose the appropriate format for each application and consult Materials and Methods sections for additional details about the use of any product in these publications.
For Research Use Only.
Not for use in diagnostic or therapeutic procedures. Not for resale. Not for distribution without written consent. Affinity Biosciences will not be held responsible for patent infringement or other violations that may occur with the use of our products. Affinity Biosciences, Affinity Biosciences Logo and all other trademarks are the property of Affinity Biosciences LTD.