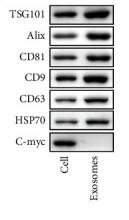
CD81 Antibody - #DF2306
| Product: | CD81 Antibody |
| Catalog: | DF2306 |
| Description: | Rabbit polyclonal antibody to CD81 |
| Application: | WB IHC IF/ICC |
| Cited expt.: | WB |
| Reactivity: | Human, Mouse, Rat |
| Prediction: | Pig, Bovine, Horse, Sheep, Dog |
| Mol.Wt.: | 26 kDa; 26kD(Calculated). |
| Uniprot: | P60033 |
| RRID: | AB_2839530 |
Related Downloads
Protocols
Product Info
*The optimal dilutions should be determined by the end user. For optimal experimental results, antibody reuse is not recommended.
*Tips:
WB: For western blot detection of denatured protein samples. IHC: For immunohistochemical detection of paraffin sections (IHC-p) or frozen sections (IHC-f) of tissue samples. IF/ICC: For immunofluorescence detection of cell samples. ELISA(peptide): For ELISA detection of antigenic peptide.
Cite Format: Affinity Biosciences Cat# DF2306, RRID:AB_2839530.
Fold/Unfold
26 kDa cell surface protein TAPA 1; 26 kDa cell surface protein TAPA-1; 26 kDa cell surface protein TAPA1; CD 81; CD81; CD81 antigen (target of antiproliferative antibody 1); CD81 antigen; CD81 molecule; CD81_HUMAN; CVID6; S5.7; TAPA 1; TAPA1; Target of the antiproliferative antibody 1; Tetraspanin 28; Tetraspanin-28; Tetraspanin28; Tspan 28; Tspan-28; Tspan28;
Immunogens
A synthesized peptide derived from human CD81, corresponding to a region within the internal amino acids.
Expressed on B cells (at protein level) (PubMed:20237408). Expressed in hepatocytes (at protein level) (PubMed:12483205). Expressed in monocytes/macrophages (at protein level) (PubMed:12796480). Expressed on both naive and memory CD4-positive T cells (at protein level) (PubMed:22307619).
- P60033 CD81_HUMAN:
- Protein BLAST With
- NCBI/
- ExPASy/
- Uniprot
MGVEGCTKCIKYLLFVFNFVFWLAGGVILGVALWLRHDPQTTNLLYLELGDKPAPNTFYVGIYILIAVGAVMMFVGFLGCYGAIQESQCLLGTFFTCLVILFACEVAAGIWGFVNKDQIAKDVKQFYDQALQQAVVDDDANNAKAVVKTFHETLDCCGSSTLTALTTSVLKNNLCPSGSNIISNLFKEDCHQKIDDLFSGKLYLIGIAAIVVAVIMIFEMILSMVLCCGIRNSSVY
Predictions
Score>80(red) has high confidence and is suggested to be used for WB detection. *The prediction model is mainly based on the alignment of immunogen sequences, the results are for reference only, not as the basis of quality assurance.
High(score>80) Medium(80>score>50) Low(score<50) No confidence
Research Backgrounds
Structural component of specialized membrane microdomains known as tetraspanin-enriched microdomains (TERMs), which act as platforms for receptor clustering and signaling. Essential for trafficking and compartmentalization of CD19 receptor on the surface of activated B cells. Upon initial encounter with microbial pathogens, enables the assembly of CD19-CR2/CD21 and B cell receptor (BCR) complexes at signaling TERMs, lowering the threshold dose of antigen required to trigger B cell clonal expansion and antibody production. In T cells, facilitates the localization of CD247/CD3 zeta at antigen-induced synapses with B cells, providing for costimulation and polarization toward T helper type 2 phenotype. Present in MHC class II compartments, may also play a role in antigen presentation. Can act both as positive and negative regulator of homotypic or heterotypic cell-cell fusion processes. Positively regulates sperm-egg fusion and may be involved in acrosome reaction (By similarity). In myoblasts, associates with CD9 and PTGFRN and inhibits myotube fusion during muscle regeneration (By similarity). In macrophages, associates with CD9 and beta-1 and beta-2 integrins, and prevents macrophage fusion into multinucleated giant cells specialized in ingesting complement-opsonized large particles. Also prevents the fusion of mononuclear cell progenitors into osteoclasts in charge of bone resorption (By similarity). May regulate the compartmentalization of enzymatic activities. In T cells, defines the subcellular localization of dNTPase SAMHD1 and permits its degradation by the proteasome, thereby controlling intracellular dNTP levels. Also involved in cell adhesion and motility. Positively regulates integrin-mediated adhesion of macrophages, particularly relevant for the inflammatory response in the lung (By similarity).
(Microbial infection) Acts as a receptor for hepatitis C virus (HCV) in hepatocytes. Association with CLDN1 and the CLDN1-CD81 receptor complex is essential for HCV entry into host cell.
(Microbial infection) Involved in SAMHD1-dependent restriction of HIV-1 replication. May support early replication of both R5- and X4-tropic HIV-1 viruses in T cells, likely via proteasome-dependent degradation of SAMHD1.
(Microbial infection) Specifically required for Plasmodium falciparum infectivity of hepatocytes, controlling sporozoite entry into hepatocytes via the parasitophorous vacuole and subsequent parasite differentiation to exoerythrocytic forms.
Not glycosylated.
Likely constitutively palmitoylated at low levels. Protein palmitoylation is up-regulated upon coligation of BCR and CD9-C2R-CD81 complexes in lipid rafts.
Cell membrane>Multi-pass membrane protein. Basolateral cell membrane>Multi-pass membrane protein.
Note: Associates with CLDN1 and the CLDN1-CD81 complex localizes to the basolateral cell membrane.
Expressed on B cells (at protein level). Expressed in hepatocytes (at protein level). Expressed in monocytes/macrophages (at protein level). Expressed on both naive and memory CD4-positive T cells (at protein level).
Binds cholesterol in a cavity lined by the transmembrane spans.
Belongs to the tetraspanin (TM4SF) family.
Research Fields
· Human Diseases > Infectious diseases: Parasitic > Malaria.
· Human Diseases > Infectious diseases: Viral > Hepatitis C.
· Organismal Systems > Immune system > B cell receptor signaling pathway. (View pathway)
References
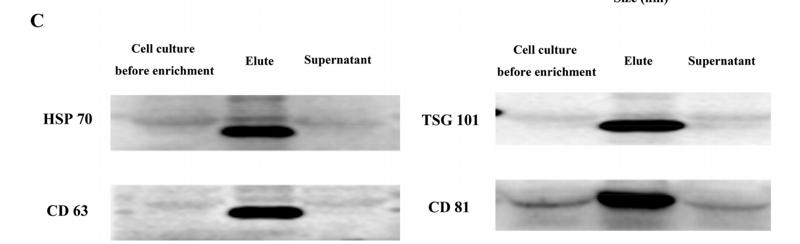
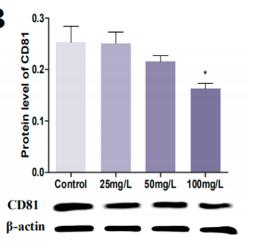
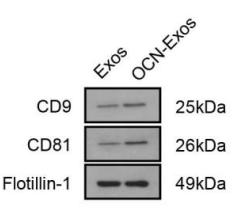
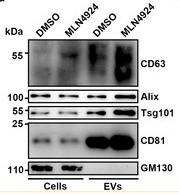
Application: WB Species: Mouse Sample:
Application: WB Species: Mice Sample: HEK293 cells
Application: WB Species: Mouse Sample:
Application: WB Species: Human Sample: H1299 cells
Application: WB Species: Mouse Sample: BM-MSCs cells
Restrictive clause
Affinity Biosciences tests all products strictly. Citations are provided as a resource for additional applications that have not been validated by Affinity Biosciences. Please choose the appropriate format for each application and consult Materials and Methods sections for additional details about the use of any product in these publications.
For Research Use Only.
Not for use in diagnostic or therapeutic procedures. Not for resale. Not for distribution without written consent. Affinity Biosciences will not be held responsible for patent infringement or other violations that may occur with the use of our products. Affinity Biosciences, Affinity Biosciences Logo and all other trademarks are the property of Affinity Biosciences LTD.