CENPF Antibody - #DF2310
| Product: | CENPF Antibody |
| Catalog: | DF2310 |
| Description: | Rabbit polyclonal antibody to CENPF |
| Application: | WB IHC |
| Cited expt.: | WB |
| Reactivity: | Human, Mouse |
| Prediction: | Pig |
| Mol.Wt.: | 367 kDa; 358kD(Calculated). |
| Uniprot: | P49454 |
| RRID: | AB_2839534 |
Related Downloads
Protocols
Product Info
*The optimal dilutions should be determined by the end user. For optimal experimental results, antibody reuse is not recommended.
*Tips:
WB: For western blot detection of denatured protein samples. IHC: For immunohistochemical detection of paraffin sections (IHC-p) or frozen sections (IHC-f) of tissue samples. IF/ICC: For immunofluorescence detection of cell samples. ELISA(peptide): For ELISA detection of antigenic peptide.
Cite Format: Affinity Biosciences Cat# DF2310, RRID:AB_2839534.
Fold/Unfold
AH antigen; Cell cycle dependent 350K nuclear protein; CENF; CENP F; CENP F kinetochore protein; CENP-F; CENPF; CENPF kinetochore protein; CENPF_HUMAN; Centromere protein F 350/400ka; Centromere protein F; Centromere protein F, 350/400kDa; CILD31; Hcp 1; Hcp1; Kinetochore protein CENP F; Kinetochore protein CENPF; Mitosin; PRO1779; STROMS;
Immunogens
A synthesized peptide derived from human CENPF, corresponding to a region within C-terminal amino acids.
- P49454 CENPF_HUMAN:
- Protein BLAST With
- NCBI/
- ExPASy/
- Uniprot
MSWALEEWKEGLPTRALQKIQELEGQLDKLKKEKQQRQFQLDSLEAALQKQKQKVENEKTEGTNLKRENQRLMEICESLEKTKQKISHELQVKESQVNFQEGQLNSGKKQIEKLEQELKRCKSELERSQQAAQSADVSLNPCNTPQKIFTTPLTPSQYYSGSKYEDLKEKYNKEVEERKRLEAEVKALQAKKASQTLPQATMNHRDIARHQASSSVFSWQQEKTPSHLSSNSQRTPIRRDFSASYFSGEQEVTPSRSTLQIGKRDANSSFFDNSSSPHLLDQLKAQNQELRNKINELELRLQGHEKEMKGQVNKFQELQLQLEKAKVELIEKEKVLNKCRDELVRTTAQYDQASTKYTALEQKLKKLTEDLSCQRQNAESARCSLEQKIKEKEKEFQEELSRQQRSFQTLDQECIQMKARLTQELQQAKNMHNVLQAELDKLTSVKQQLENNLEEFKQKLCRAEQAFQASQIKENELRRSMEEMKKENNLLKSHSEQKAREVCHLEAELKNIKQCLNQSQNFAEEMKAKNTSQETMLRDLQEKINQQENSLTLEKLKLAVADLEKQRDCSQDLLKKREHHIEQLNDKLSKTEKESKALLSALELKKKEYEELKEEKTLFSCWKSENEKLLTQMESEKENLQSKINHLETCLKTQQIKSHEYNERVRTLEMDRENLSVEIRNLHNVLDSKSVEVETQKLAYMELQQKAEFSDQKHQKEIENMCLKTSQLTGQVEDLEHKLQLLSNEIMDKDRCYQDLHAEYESLRDLLKSKDASLVTNEDHQRSLLAFDQQPAMHHSFANIIGEQGSMPSERSECRLEADQSPKNSAILQNRVDSLEFSLESQKQMNSDLQKQCEELVQIKGEIEENLMKAEQMHQSFVAETSQRISKLQEDTSAHQNVVAETLSALENKEKELQLLNDKVETEQAEIQELKKSNHLLEDSLKELQLLSETLSLEKKEMSSIISLNKREIEELTQENGTLKEINASLNQEKMNLIQKSESFANYIDEREKSISELSDQYKQEKLILLQRCEETGNAYEDLSQKYKAAQEKNSKLECLLNECTSLCENRKNELEQLKEAFAKEHQEFLTKLAFAEERNQNLMLELETVQQALRSEMTDNQNNSKSEAGGLKQEIMTLKEEQNKMQKEVNDLLQENEQLMKVMKTKHECQNLESEPIRNSVKERESERNQCNFKPQMDLEVKEISLDSYNAQLVQLEAMLRNKELKLQESEKEKECLQHELQTIRGDLETSNLQDMQSQEISGLKDCEIDAEEKYISGPHELSTSQNDNAHLQCSLQTTMNKLNELEKICEILQAEKYELVTELNDSRSECITATRKMAEEVGKLLNEVKILNDDSGLLHGELVEDIPGGEFGEQPNEQHPVSLAPLDESNSYEHLTLSDKEVQMHFAELQEKFLSLQSEHKILHDQHCQMSSKMSELQTYVDSLKAENLVLSTNLRNFQGDLVKEMQLGLEEGLVPSLSSSCVPDSSSLSSLGDSSFYRALLEQTGDMSLLSNLEGAVSANQCSVDEVFCSSLQEENLTRKETPSAPAKGVEELESLCEVYRQSLEKLEEKMESQGIMKNKEIQELEQLLSSERQELDCLRKQYLSENEQWQQKLTSVTLEMESKLAAEKKQTEQLSLELEVARLQLQGLDLSSRSLLGIDTEDAIQGRNESCDISKEHTSETTERTPKHDVHQICDKDAQQDLNLDIEKITETGAVKPTGECSGEQSPDTNYEPPGEDKTQGSSECISELSFSGPNALVPMDFLGNQEDIHNLQLRVKETSNENLRLLHVIEDRDRKVESLLNEMKELDSKLHLQEVQLMTKIEACIELEKIVGELKKENSDLSEKLEYFSCDHQELLQRVETSEGLNSDLEMHADKSSREDIGDNVAKVNDSWKERFLDVENELSRIRSEKASIEHEALYLEADLEVVQTEKLCLEKDNENKQKVIVCLEEELSVVTSERNQLRGELDTMSKKTTALDQLSEKMKEKTQELESHQSECLHCIQVAEAEVKEKTELLQTLSSDVSELLKDKTHLQEKLQSLEKDSQALSLTKCELENQIAQLNKEKELLVKESESLQARLSESDYEKLNVSKALEAALVEKGEFALRLSSTQEEVHQLRRGIEKLRVRIEADEKKQLHIAEKLKERERENDSLKDKVENLERELQMSEENQELVILDAENSKAEVETLKTQIEEMARSLKVFELDLVTLRSEKENLTKQIQEKQGQLSELDKLLSSFKSLLEEKEQAEIQIKEESKTAVEMLQNQLKELNEAVAALCGDQEIMKATEQSLDPPIEEEHQLRNSIEKLRARLEADEKKQLCVLQQLKESEHHADLLKGRVENLERELEIARTNQEHAALEAENSKGEVETLKAKIEGMTQSLRGLELDVVTIRSEKENLTNELQKEQERISELEIINSSFENILQEKEQEKVQMKEKSSTAMEMLQTQLKELNERVAALHNDQEACKAKEQNLSSQVECLELEKAQLLQGLDEAKNNYIVLQSSVNGLIQEVEDGKQKLEKKDEEISRLKNQIQDQEQLVSKLSQVEGEHQLWKEQNLELRNLTVELEQKIQVLQSKNASLQDTLEVLQSSYKNLENELELTKMDKMSFVEKVNKMTAKETELQREMHEMAQKTAELQEELSGEKNRLAGELQLLLEEIKSSKDQLKELTLENSELKKSLDCMHKDQVEKEGKVREEIAEYQLRLHEAEKKHQALLLDTNKQYEVEIQTYREKLTSKEECLSSQKLEIDLLKSSKEELNNSLKATTQILEELKKTKMDNLKYVNQLKKENERAQGKMKLLIKSCKQLEEEKEILQKELSQLQAAQEKQKTGTVMDTKVDELTTEIKELKETLEEKTKEADEYLDKYCSLLISHEKLEKAKEMLETQVAHLCSQQSKQDSRGSPLLGPVVPGPSPIPSVTEKRLSSGQNKASGKRQRSSGIWENGRGPTPATPESFSKKSKKAVMSGIHPAEDTEGTEFEPEGLPEVVKKGFADIPTGKTSPYILRRTTMATRTSPRLAAQKLALSPLSLGKENLAESSKPTAGGSRSQKVKVAQRSPVDSGTILREPTTKSVPVNNLPERSPTDSPREGLRVKRGRLVPSPKAGLESNGSENCKVQ
Predictions
Score>80(red) has high confidence and is suggested to be used for WB detection. *The prediction model is mainly based on the alignment of immunogen sequences, the results are for reference only, not as the basis of quality assurance.
High(score>80) Medium(80>score>50) Low(score<50) No confidence
Research Backgrounds
Required for kinetochore function and chromosome segregation in mitosis. Required for kinetochore localization of dynein, LIS1, NDE1 and NDEL1. Regulates recycling of the plasma membrane by acting as a link between recycling vesicles and the microtubule network though its association with STX4 and SNAP25. Acts as a potential inhibitor of pocket protein-mediated cellular processes during development by regulating the activity of RB proteins during cell division and proliferation. May play a regulatory or permissive role in the normal embryonic cardiomyocyte cell cycle and in promoting continued mitosis in transformed, abnormally dividing neonatal cardiomyocytes. Interaction with RB directs embryonic stem cells toward a cardiac lineage. Involved in the regulation of DNA synthesis and hence cell cycle progression, via its C-terminus. Has a potential role regulating skeletal myogenesis and in cell differentiation in embryogenesis. Involved in dendritic cell regulation of T-cell immunity against chlamydia.
Hyperphosphorylated during mitosis.
Cytoplasm>Perinuclear region. Nucleus matrix. Chromosome>Centromere>Kinetochore. Cytoplasm>Cytoskeleton>Spindle.
Note: Relocalizes to the kinetochore/centromere (coronal surface of the outer plate) and the spindle during mitosis. Observed in nucleus during interphase but not in the nucleolus. At metaphase becomes localized to areas including kinetochore and mitotic apparatus as well as cytoplasm. By telophase, is concentrated within the intracellular bridge at either side of the mid-body.
Belongs to the centromere protein F family.
References
Application: WB Species: human Sample: MCF-7, T47D, MDA-MB-231 and MDA-MB-231/ADR cells
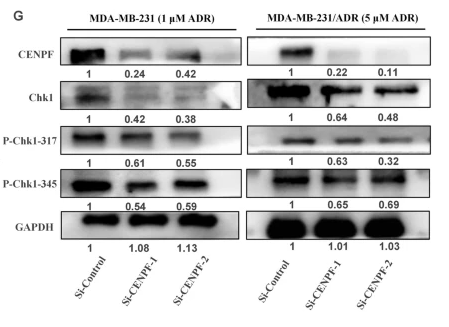
Application: WB Species: Human Sample: MDA-MB-231 and MDA-MB-231/ADR cells
Restrictive clause
Affinity Biosciences tests all products strictly. Citations are provided as a resource for additional applications that have not been validated by Affinity Biosciences. Please choose the appropriate format for each application and consult Materials and Methods sections for additional details about the use of any product in these publications.
For Research Use Only.
Not for use in diagnostic or therapeutic procedures. Not for resale. Not for distribution without written consent. Affinity Biosciences will not be held responsible for patent infringement or other violations that may occur with the use of our products. Affinity Biosciences, Affinity Biosciences Logo and all other trademarks are the property of Affinity Biosciences LTD.