Phospho-FOXO3A (Ser253) Antibody - #AF3020
| Product: | Phospho-FOXO3A (Ser253) Antibody |
| Catalog: | AF3020 |
| Description: | Rabbit polyclonal antibody to Phospho-FOXO3A (Ser253) |
| Application: | WB IHC IF/ICC |
| Cited expt.: | WB |
| Reactivity: | Human, Mouse, Rat |
| Prediction: | Pig, Bovine, Horse, Sheep, Rabbit, Dog, Chicken, Xenopus |
| Mol.Wt.: | 97kDa; 71kD(Calculated). |
| Uniprot: | O43524 |
| RRID: | AB_2834427 |
Product Info
*The optimal dilutions should be determined by the end user. For optimal experimental results, antibody reuse is not recommended.
*Tips:
WB: For western blot detection of denatured protein samples. IHC: For immunohistochemical detection of paraffin sections (IHC-p) or frozen sections (IHC-f) of tissue samples. IF/ICC: For immunofluorescence detection of cell samples. ELISA(peptide): For ELISA detection of antigenic peptide.
Cite Format: Affinity Biosciences Cat# AF3020, RRID:AB_2834427.
Fold/Unfold
AF6q21; AF6q21 protein; DKFZp781A0677; FKHR2; FKHRL 1; FKHRL1; FKHRL1P2; Forkhead (Drosophila) homolog (rhabdomyosarcoma) like 1; Forkhead box O3; Forkhead box O3A; Forkhead box protein O3; Forkhead box protein O3A; Forkhead Drosophila homolog of in rhabdomyosarcoma like 1; Forkhead homolog (rhabdomyosarcoma) like 1; Forkhead in rhabdomyosarcoma like 1; Forkhead in rhabdomyosarcoma-like 1; FOX O3A; FOXO2; foxo3; FOXO3_HUMAN; FOXO3A; MGC12739; MGC31925;
Immunogens
A synthesized peptide derived from human FOXO3A around the phosphorylation site of Ser253.
- O43524 FOXO3_HUMAN:
- Protein BLAST With
- NCBI/
- ExPASy/
- Uniprot
MAEAPASPAPLSPLEVELDPEFEPQSRPRSCTWPLQRPELQASPAKPSGETAADSMIPEEEDDEDDEDGGGRAGSAMAIGGGGGSGTLGSGLLLEDSARVLAPGGQDPGSGPATAAGGLSGGTQALLQPQQPLPPPQPGAAGGSGQPRKCSSRRNAWGNLSYADLITRAIESSPDKRLTLSQIYEWMVRCVPYFKDKGDSNSSAGWKNSIRHNLSLHSRFMRVQNEGTGKSSWWIINPDGGKSGKAPRRRAVSMDNSNKYTKSRGRAAKKKAALQTAPESADDSPSQLSKWPGSPTSRSSDELDAWTDFRSRTNSNASTVSGRLSPIMASTELDEVQDDDAPLSPMLYSSSASLSPSVSKPCTVELPRLTDMAGTMNLNDGLTENLMDDLLDNITLPPSQPSPTGGLMQRSSSFPYTTKGSGLGSPTSSFNSTVFGPSSLNSLRQSPMQTIQENKPATFSSMSHYGNQTLQDLLTSDSLSHSDVMMTQSDPLMSQASTAVSAQNSRRNVMLRNDPMMSFAAQPNQGSLVNQNLLHHQHQTQGALGGSRALSNSVSNMGLSESSSLGSAKHQQQSPVSQSMQTLSDSLSGSSLYSTSANLPVMGHEKFPSDLDLDMFNGSLECDMESIIRSELMDADGLDFNFDSLISTQNVVGLNVGNFTGAKQASSQSWVPG
Predictions
Score>80(red) has high confidence and is suggested to be used for WB detection. *The prediction model is mainly based on the alignment of immunogen sequences, the results are for reference only, not as the basis of quality assurance.
High(score>80) Medium(80>score>50) Low(score<50) No confidence
Research Backgrounds
Transcriptional activator that recognizes and binds to the DNA sequence 5'-[AG]TAAA[TC]A-3' and regulates different processes, such as apoptosis and autophagy. Acts as a positive regulator of autophagy in skeletal muscle: in starved cells, enters the nucleus following dephosphorylation and binds the promoters of autophagy genes, such as GABARAP1L, MAP1LC3B and ATG12, thereby activating their expression, resulting in proteolysis of skeletal muscle proteins (By similarity). Triggers apoptosis in the absence of survival factors, including neuronal cell death upon oxidative stress. Participates in post-transcriptional regulation of MYC: following phosphorylation by MAPKAPK5, promotes induction of miR-34b and miR-34c expression, 2 post-transcriptional regulators of MYC that bind to the 3'UTR of MYC transcript and prevent its translation. In response to metabolic stress, translocates into the mitochondria where it promotes mtDNA transcription.
In the presence of survival factors such as IGF-1, phosphorylated on Thr-32 and Ser-253 by AKT1/PKB. This phosphorylated form then interacts with 14-3-3 proteins and is retained in the cytoplasm. Survival factor withdrawal induces dephosphorylation and promotes translocation to the nucleus where the dephosphorylated protein induces transcription of target genes and triggers apoptosis. Although AKT1/PKB doesn't appear to phosphorylate Ser-315 directly, it may activate other kinases that trigger phosphorylation at this residue. Phosphorylated by STK4/MST1 on Ser-209 upon oxidative stress, which leads to dissociation from YWHAB/14-3-3-beta and nuclear translocation. Phosphorylated by PIM1. Phosphorylation by AMPK leads to the activation of transcriptional activity without affecting subcellular localization. In response to metabolic stress, phosphorylated by AMPK on Ser-30 which mediates FOXO3 mitochondrial translocation. Phosphorylation by MAPKAPK5 promotes nuclear localization and DNA-binding, leading to induction of miR-34b and miR-34c expression, 2 post-transcriptional regulators of MYC that bind to the 3'UTR of MYC transcript and prevent its translation. Phosphorylated by CHUK/IKKA and IKBKB/IKKB. TNF-induced inactivation of FOXO3 requires its phosphorylation at Ser-644 by IKBKB/IKKB which promotes FOXO3 retention in the cytoplasm, polyubiquitination and ubiquitin-mediated proteasomal degradation. May be dephosphorylated by calcineurin A on Ser-299 which abolishes FOXO3 transcriptional activity (By similarity). In cancer cells, ERK mediated-phosphorylation of Ser-12 is required for mitochondrial translocation of FOXO3 in response to metabolic stress or chemotherapeutic agents.
Deacetylation by SIRT1 or SIRT2 stimulates interaction of FOXO3 with SKP2 and facilitates SCF(SKP2)-mediated FOXO3 ubiquitination and proteasomal degradation. Deacetylation by SIRT2 stimulates FOXO3-mediated transcriptional activity in response to oxidative stress (By similarity). Deacetylated by SIRT3. Deacetylation by SIRT3 stimulates FOXO3-mediated mtDNA transcriptional activity in response to metabolic stress.
Heavily methylated by SET9 which decreases stability, while moderately increasing transcriptional activity. The main methylation site is Lys-271. Methylation doesn't affect subcellular location.
Polyubiquitinated. Ubiquitinated by a SCF complex containing SKP2, leading to proteasomal degradation.
The N-terminus is cleaved following import into the mitochondrion.
Cytoplasm>Cytosol. Nucleus. Mitochondrion matrix. Mitochondrion outer membrane>Peripheral membrane protein>Cytoplasmic side.
Note: Retention in the cytoplasm contributes to its inactivation (PubMed:10102273, PubMed:15084260, PubMed:16751106). Translocates to the nucleus upon oxidative stress and in the absence of survival factors (PubMed:10102273, PubMed:16751106). Translocates from the cytosol to the nucleus following dephosphorylation in response to autophagy-inducing stimuli (By similarity). Translocates in a AMPK-dependent manner into the mitochondrion in response to metabolic stress (PubMed:23283301, PubMed:29445193).
Ubiquitous.
Research Fields
· Cellular Processes > Cell growth and death > Cellular senescence. (View pathway)
· Environmental Information Processing > Signal transduction > FoxO signaling pathway. (View pathway)
· Environmental Information Processing > Signal transduction > PI3K-Akt signaling pathway. (View pathway)
· Environmental Information Processing > Signal transduction > AMPK signaling pathway. (View pathway)
· Human Diseases > Drug resistance: Antineoplastic > EGFR tyrosine kinase inhibitor resistance.
· Human Diseases > Cancers: Specific types > Endometrial cancer. (View pathway)
· Human Diseases > Cancers: Specific types > Non-small cell lung cancer. (View pathway)
· Organismal Systems > Immune system > Chemokine signaling pathway. (View pathway)
· Organismal Systems > Aging > Longevity regulating pathway. (View pathway)
· Organismal Systems > Aging > Longevity regulating pathway - multiple species. (View pathway)
· Organismal Systems > Nervous system > Neurotrophin signaling pathway. (View pathway)
· Organismal Systems > Endocrine system > Prolactin signaling pathway. (View pathway)
References
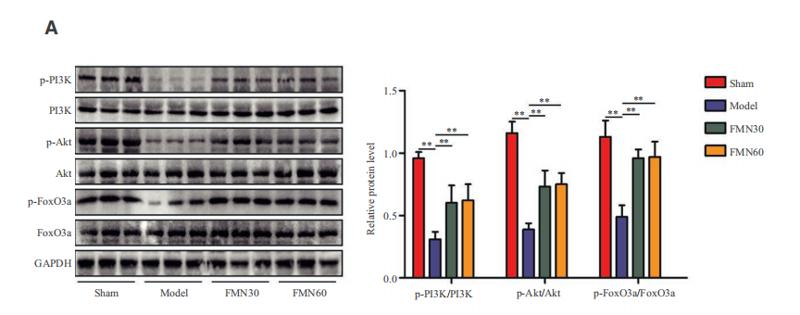
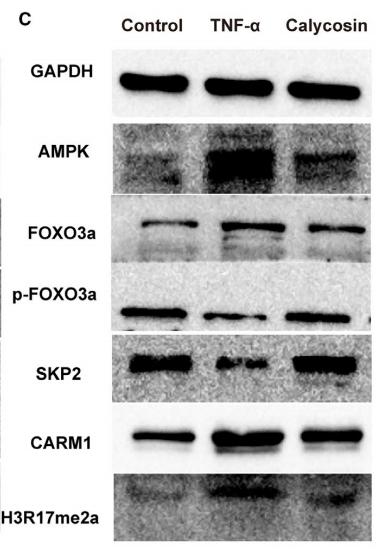
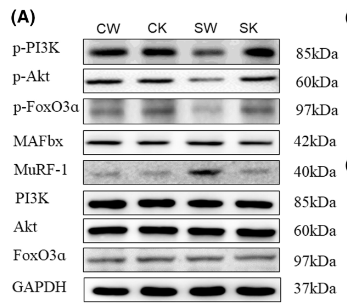
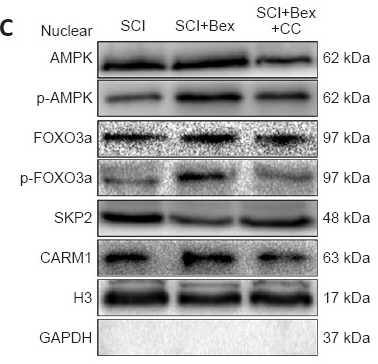
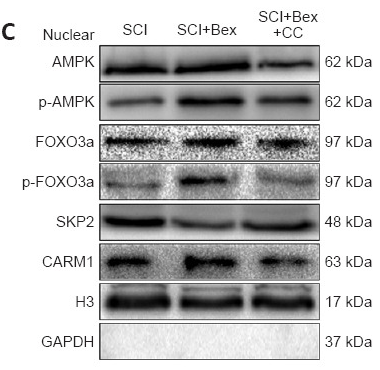
Application: WB Species: Mouse Sample: spinal cord
Application: WB Species: Mouse Sample: spinal cord
Application: WB Species: Mouse Sample: breast cancer cells
Application: WB Species: Mouse Sample: C2C12 myoblasts
Application: WB Species: mouse Sample: C2C12 cells
Application: WB Species: Rat Sample: spinal cord
Restrictive clause
Affinity Biosciences tests all products strictly. Citations are provided as a resource for additional applications that have not been validated by Affinity Biosciences. Please choose the appropriate format for each application and consult Materials and Methods sections for additional details about the use of any product in these publications.
For Research Use Only.
Not for use in diagnostic or therapeutic procedures. Not for resale. Not for distribution without written consent. Affinity Biosciences will not be held responsible for patent infringement or other violations that may occur with the use of our products. Affinity Biosciences, Affinity Biosciences Logo and all other trademarks are the property of Affinity Biosciences LTD.