Phospho-STAT1 (Tyr701) Antibody - #AF3300
| Product: | Phospho-STAT1 (Tyr701) Antibody |
| Catalog: | AF3300 |
| Description: | Rabbit polyclonal antibody to Phospho-STAT1 (Tyr701) |
| Application: | WB IHC IF/ICC IP |
| Cited expt.: | WB, IHC, IF/ICC |
| Reactivity: | Human, Mouse, Rat |
| Prediction: | Pig, Bovine, Horse, Rabbit, Dog, Chicken, Xenopus |
| Mol.Wt.: | 84kDa; 87kD(Calculated). |
| Uniprot: | P42224 |
| RRID: | AB_2834719 |
Product Info
*The optimal dilutions should be determined by the end user. For optimal experimental results, antibody reuse is not recommended.
*Tips:
WB: For western blot detection of denatured protein samples. IHC: For immunohistochemical detection of paraffin sections (IHC-p) or frozen sections (IHC-f) of tissue samples. IF/ICC: For immunofluorescence detection of cell samples. ELISA(peptide): For ELISA detection of antigenic peptide.
Cite Format: Affinity Biosciences Cat# AF3300, RRID:AB_2834719.
Fold/Unfold
CANDF7; DKFZp686B04100; ISGF 3; ISGF3; OTTHUMP00000163552; OTTHUMP00000165046; OTTHUMP00000165047; OTTHUMP00000205845; Signal transducer and activator of transcription 1; Signal transducer and activator of transcription 1, 91kDa; Signal transducer and activator of transcription 1-alpha/beta; Stat1; STAT1_HUMAN; STAT91; Transcription factor ISGF-3 components p91/p84;
Immunogens
A synthesized peptide derived from human STAT1 around the phosphorylation site of Tyr701.
- P42224 STAT1_HUMAN:
- Protein BLAST With
- NCBI/
- ExPASy/
- Uniprot
MSQWYELQQLDSKFLEQVHQLYDDSFPMEIRQYLAQWLEKQDWEHAANDVSFATIRFHDLLSQLDDQYSRFSLENNFLLQHNIRKSKRNLQDNFQEDPIQMSMIIYSCLKEERKILENAQRFNQAQSGNIQSTVMLDKQKELDSKVRNVKDKVMCIEHEIKSLEDLQDEYDFKCKTLQNREHETNGVAKSDQKQEQLLLKKMYLMLDNKRKEVVHKIIELLNVTELTQNALINDELVEWKRRQQSACIGGPPNACLDQLQNWFTIVAESLQQVRQQLKKLEELEQKYTYEHDPITKNKQVLWDRTFSLFQQLIQSSFVVERQPCMPTHPQRPLVLKTGVQFTVKLRLLVKLQELNYNLKVKVLFDKDVNERNTVKGFRKFNILGTHTKVMNMEESTNGSLAAEFRHLQLKEQKNAGTRTNEGPLIVTEELHSLSFETQLCQPGLVIDLETTSLPVVVISNVSQLPSGWASILWYNMLVAEPRNLSFFLTPPCARWAQLSEVLSWQFSSVTKRGLNVDQLNMLGEKLLGPNASPDGLIPWTRFCKENINDKNFPFWLWIESILELIKKHLLPLWNDGCIMGFISKERERALLKDQQPGTFLLRFSESSREGAITFTWVERSQNGGEPDFHAVEPYTKKELSAVTFPDIIRNYKVMAAENIPENPLKYLYPNIDKDHAFGKYYSRPKEAPEPMELDGPKGTGYIKTELISVSEVHPSRLQTTDNLLPMSPEEFDEVSRIVGSVEFDSMMNTV
Predictions
Score>80(red) has high confidence and is suggested to be used for WB detection. *The prediction model is mainly based on the alignment of immunogen sequences, the results are for reference only, not as the basis of quality assurance.
High(score>80) Medium(80>score>50) Low(score<50) No confidence
Research Backgrounds
Signal transducer and transcription activator that mediates cellular responses to interferons (IFNs), cytokine KITLG/SCF and other cytokines and other growth factors. Following type I IFN (IFN-alpha and IFN-beta) binding to cell surface receptors, signaling via protein kinases leads to activation of Jak kinases (TYK2 and JAK1) and to tyrosine phosphorylation of STAT1 and STAT2. The phosphorylated STATs dimerize and associate with ISGF3G/IRF-9 to form a complex termed ISGF3 transcription factor, that enters the nucleus. ISGF3 binds to the IFN stimulated response element (ISRE) to activate the transcription of IFN-stimulated genes (ISG), which drive the cell in an antiviral state. In response to type II IFN (IFN-gamma), STAT1 is tyrosine- and serine-phosphorylated. It then forms a homodimer termed IFN-gamma-activated factor (GAF), migrates into the nucleus and binds to the IFN gamma activated sequence (GAS) to drive the expression of the target genes, inducing a cellular antiviral state. Becomes activated in response to KITLG/SCF and KIT signaling. May mediate cellular responses to activated FGFR1, FGFR2, FGFR3 and FGFR4.
Phosphorylated on tyrosine and serine residues in response to a variety of cytokines/growth hormones including IFN-alpha, IFN-gamma, PDGF and EGF. Activated KIT promotes phosphorylation on tyrosine residues and subsequent translocation to the nucleus. Upon EGF stimulation, phosphorylation on Tyr-701 (lacking in beta form) by JAK1, JAK2 or TYK2 promotes dimerization and subsequent translocation to the nucleus. Growth hormone (GH) activates STAT1 signaling only via JAK2. Tyrosine phosphorylated in response to constitutively activated FGFR1, FGFR2, FGFR3 and FGFR4. Phosphorylation on Ser-727 by several kinases including MAPK14, ERK1/2 and CAMKII on IFN-gamma stimulation, regulates STAT1 transcriptional activity. Phosphorylation on Ser-727 promotes sumoylation though increasing interaction with PIAS. Phosphorylation on Ser-727 by PRKCD induces apoptosis in response to DNA-damaging agents. Phosphorylated on tyrosine residues when PTK2/FAK1 is activated; most likely this is catalyzed by a SRC family kinase. Dephosphorylation on tyrosine residues by PTPN2 negatively regulates interferon-mediated signaling. Upon viral infection or IFN induction, phosphorylation on Ser-708 occurs much later than phosphorylation on Tyr-701 and is required for the binding of ISGF3 on the ISREs of a subset of IFN-stimulated genes IKBKE-dependent. Phosphorylation at Tyr-701 and Ser-708 are mutually exclusive, phosphorylation at Ser-708 requires previous dephosphorylation of Tyr-701.
Sumoylated with SUMO1, SUMO2 and SUMO3. Sumoylation is enhanced by IFN-gamma-induced phosphorylation on Ser-727, and by interaction with PIAS proteins. Enhances the transactivation activity.
ISGylated.
Mono-ADP-ribosylated at Glu-657 and Glu-705 by PARP14; ADP-ribosylation prevents phosphorylation at Tyr-701. However, the role of ADP-ribosylation in the prevention of phosphorylation has been called into question and the lack of phosphorylation may be due to sumoylation of Lys-703.
Monomethylated at Lys-525 by SETD2; monomethylation is necessary for phosphorylation at Tyr-701, translocation into the nucleus and activation of the antiviral defense.
Cytoplasm. Nucleus.
Note: Translocated into the nucleus upon tyrosine phosphorylation and dimerization, in response to IFN-gamma and signaling by activated FGFR1, FGFR2, FGFR3 or FGFR4 (PubMed:15322115). Monomethylation at Lys-525 is required for phosphorylation at Tyr-701 and translocation into the nucleus (PubMed:28753426). Translocates into the nucleus in response to interferon-beta stimulation (PubMed:26479788).
Belongs to the transcription factor STAT family.
Research Fields
· Cellular Processes > Cell growth and death > Necroptosis. (View pathway)
· Environmental Information Processing > Signal transduction > Jak-STAT signaling pathway. (View pathway)
· Human Diseases > Infectious diseases: Parasitic > Leishmaniasis.
· Human Diseases > Infectious diseases: Parasitic > Toxoplasmosis.
· Human Diseases > Infectious diseases: Bacterial > Tuberculosis.
· Human Diseases > Infectious diseases: Viral > Hepatitis C.
· Human Diseases > Infectious diseases: Viral > Hepatitis B.
· Human Diseases > Infectious diseases: Viral > Measles.
· Human Diseases > Infectious diseases: Viral > Influenza A.
· Human Diseases > Infectious diseases: Viral > Human papillomavirus infection.
· Human Diseases > Infectious diseases: Viral > Herpes simplex infection.
· Human Diseases > Cancers: Overview > Pathways in cancer. (View pathway)
· Human Diseases > Cancers: Specific types > Pancreatic cancer. (View pathway)
· Human Diseases > Immune diseases > Inflammatory bowel disease (IBD).
· Organismal Systems > Immune system > Chemokine signaling pathway. (View pathway)
· Organismal Systems > Development > Osteoclast differentiation. (View pathway)
· Organismal Systems > Immune system > Toll-like receptor signaling pathway. (View pathway)
· Organismal Systems > Immune system > NOD-like receptor signaling pathway. (View pathway)
· Organismal Systems > Immune system > Th1 and Th2 cell differentiation. (View pathway)
· Organismal Systems > Immune system > Th17 cell differentiation. (View pathway)
· Organismal Systems > Endocrine system > Prolactin signaling pathway. (View pathway)
· Organismal Systems > Endocrine system > Thyroid hormone signaling pathway. (View pathway)
References
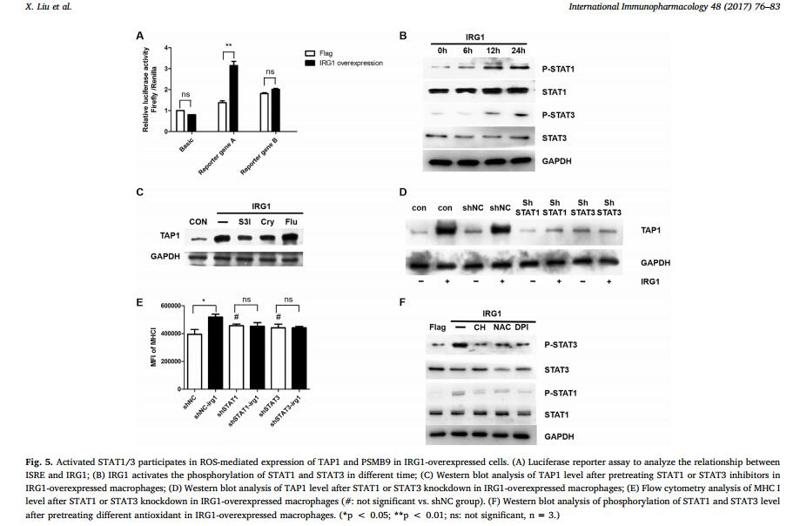
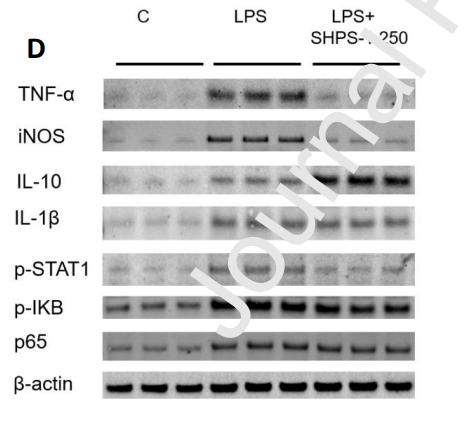
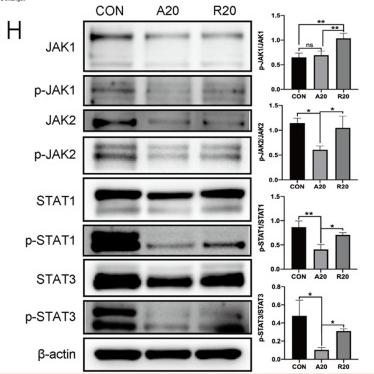
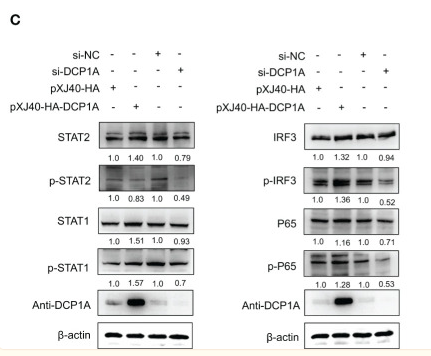
Application: WB Species: Mouse Sample: RAW 264.7 cells
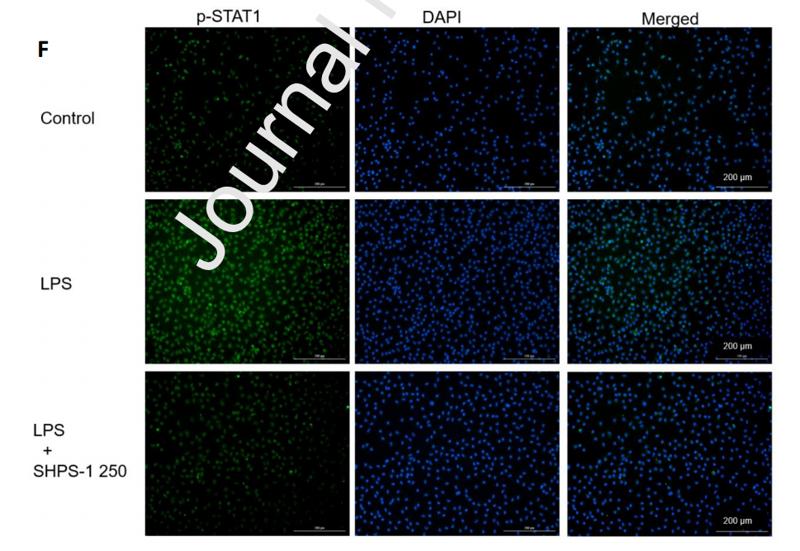
Application: IF/ICC Species: Mouse Sample: RAW 264.7 cells
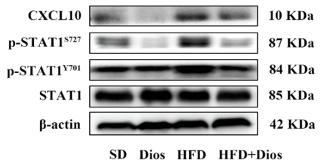
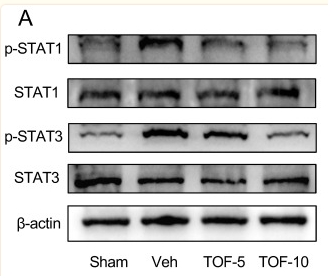
Application: WB Species: Mice Sample:
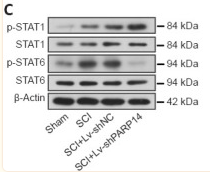
Application: WB Species: Rat Sample: spinal cord
Restrictive clause
Affinity Biosciences tests all products strictly. Citations are provided as a resource for additional applications that have not been validated by Affinity Biosciences. Please choose the appropriate format for each application and consult Materials and Methods sections for additional details about the use of any product in these publications.
For Research Use Only.
Not for use in diagnostic or therapeutic procedures. Not for resale. Not for distribution without written consent. Affinity Biosciences will not be held responsible for patent infringement or other violations that may occur with the use of our products. Affinity Biosciences, Affinity Biosciences Logo and all other trademarks are the property of Affinity Biosciences LTD.