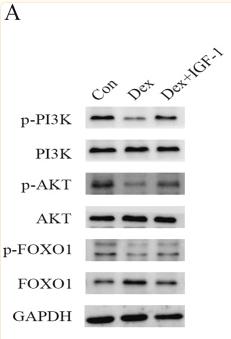
Phospho-FOXO1A (Ser256) Antibody - #AF3417
| Product: | Phospho-FOXO1A (Ser256) Antibody |
| Catalog: | AF3417 |
| Description: | Rabbit polyclonal antibody to Phospho-FOXO1A (Ser256) |
| Application: | WB IHC IF/ICC |
| Cited expt.: | WB, IHC |
| Reactivity: | Human, Mouse, Rat |
| Prediction: | Pig, Bovine, Dog, Chicken, Xenopus |
| Mol.Wt.: | 69kDa; 70kD(Calculated). |
| Uniprot: | Q12778 |
| RRID: | AB_2834859 |
Product Info
*The optimal dilutions should be determined by the end user. For optimal experimental results, antibody reuse is not recommended.
*Tips:
WB: For western blot detection of denatured protein samples. IHC: For immunohistochemical detection of paraffin sections (IHC-p) or frozen sections (IHC-f) of tissue samples. IF/ICC: For immunofluorescence detection of cell samples. ELISA(peptide): For ELISA detection of antigenic peptide.
Cite Format: Affinity Biosciences Cat# AF3417, RRID:AB_2834859.
Fold/Unfold
FKH 1; FKH1; FKHR; Forkhead (Drosophila) homolog 1 (rhabdomyosarcoma); Forkhead box O1; Forkhead box protein O1; Forkhead box protein O1A; Forkhead in rhabdomyosarcoma; Forkhead, Drosophila, homolog of, in rhabdomyosarcoma; FoxO transcription factor; foxo1; FOXO1_HUMAN; FOXO1A; OTTHUMP00000018301;
Immunogens
A synthesized peptide derived from human FOXO1A around the phosphorylation site of Ser256.
- Q12778 FOXO1_HUMAN:
- Protein BLAST With
- NCBI/
- ExPASy/
- Uniprot
MAEAPQVVEIDPDFEPLPRPRSCTWPLPRPEFSQSNSATSSPAPSGSAAANPDAAAGLPSASAAAVSADFMSNLSLLEESEDFPQAPGSVAAAVAAAAAAAATGGLCGDFQGPEAGCLHPAPPQPPPPGPLSQHPPVPPAAAGPLAGQPRKSSSSRRNAWGNLSYADLITKAIESSAEKRLTLSQIYEWMVKSVPYFKDKGDSNSSAGWKNSIRHNLSLHSKFIRVQNEGTGKSSWWMLNPEGGKSGKSPRRRAASMDNNSKFAKSRSRAAKKKASLQSGQEGAGDSPGSQFSKWPASPGSHSNDDFDNWSTFRPRTSSNASTISGRLSPIMTEQDDLGEGDVHSMVYPPSAAKMASTLPSLSEISNPENMENLLDNLNLLSSPTSLTVSTQSSPGTMMQQTPCYSFAPPNTSLNSPSPNYQKYTYGQSSMSPLPQMPIQTLQDNKSSYGGMSQYNCAPGLLKELLTSDSPPHNDIMTPVDPGVAQPNSRVLGQNVMMGPNSVMSTYGSQASHNKMMNPSSHTHPGHAQQTSAVNGRPLPHTVSTMPHTSGMNRLTQVKTPVQVPLPHPMQMSALGGYSSVSSCNGYGRMGLLHQEKLPSDLDGMFIERLDCDMESIIRNDLMDGDTLDFNFDNVLPNQSFPHSVKTTTHSWVSG
Predictions
Score>80(red) has high confidence and is suggested to be used for WB detection. *The prediction model is mainly based on the alignment of immunogen sequences, the results are for reference only, not as the basis of quality assurance.
High(score>80) Medium(80>score>50) Low(score<50) No confidence
Research Backgrounds
Transcription factor that is the main target of insulin signaling and regulates metabolic homeostasis in response to oxidative stress. Binds to the insulin response element (IRE) with consensus sequence 5'-TT[G/A]TTTTG-3' and the related Daf-16 family binding element (DBE) with consensus sequence 5'-TT[G/A]TTTAC-3'. Activity suppressed by insulin. Main regulator of redox balance and osteoblast numbers and controls bone mass. Orchestrates the endocrine function of the skeleton in regulating glucose metabolism. Acts synergistically with ATF4 to suppress osteocalcin/BGLAP activity, increasing glucose levels and triggering glucose intolerance and insulin insensitivity. Also suppresses the transcriptional activity of RUNX2, an upstream activator of osteocalcin/BGLAP. In hepatocytes, promotes gluconeogenesis by acting together with PPARGC1A and CEBPA to activate the expression of genes such as IGFBP1, G6PC and PCK1. Important regulator of cell death acting downstream of CDK1, PKB/AKT1 and STK4/MST1. Promotes neural cell death. Mediates insulin action on adipose tissue. Regulates the expression of adipogenic genes such as PPARG during preadipocyte differentiation and, adipocyte size and adipose tissue-specific gene expression in response to excessive calorie intake. Regulates the transcriptional activity of GADD45A and repair of nitric oxide-damaged DNA in beta-cells. Required for the autophagic cell death induction in response to starvation or oxidative stress in a transcription-independent manner. Mediates the function of MLIP in cardiomyocytes hypertrophy and cardiac remodeling (By similarity).
Phosphorylation by NLK promotes nuclear export and inhibits the transcriptional activity. In response to growth factors, phosphorylation on Thr-24, Ser-256 and Ser-322 by PKB/AKT1 promotes nuclear export and inactivation of transactivational activity. Phosphorylation on Thr-24 is required for binding 14-3-3 proteins. Phosphorylation of Ser-256 decreases DNA-binding activity and promotes the phosphorylation of Thr-24 and Ser-319, permitting phosphorylation of Ser-322 and Ser-325, probably by CDK1, leading to nuclear exclusion and loss of function. Stress signals, such as response to oxygen or nitric oxide, attenuate the PKB/AKT1-mediated phosphorylation leading to nuclear retention. Phosphorylation of Ser-329 is independent of IGF1 and leads to reduced function. Dephosphorylated on Thr-24 and Ser-256 by PP2A in beta-cells under oxidative stress leading to nuclear retention (By similarity). Phosphorylation of Ser-249 by CDK1 disrupts binding of 14-3-3 proteins leading to nuclear accumulation and has no effect on DNA-binding nor transcriptional activity. Phosphorylation by STK4/MST1 on Ser-212, upon oxidative stress, inhibits binding to 14-3-3 proteins and nuclear export.
Acetylated. Acetylation at Lys-262, Lys-265 and Lys-274 are necessary for autophagic cell death induction. Deacetylated by SIRT2 in response to oxidative stress or serum deprivation, thereby negatively regulating FOXO1-mediated autophagic cell death.
Ubiquitinated by SKP2. Ubiquitination leads to proteasomal degradation.
Methylation inhibits AKT1-mediated phosphorylation at Ser-256 and is increased by oxidative stress.
Once in the nucleus, acetylated by CREBBP/EP300. Acetylation diminishes the interaction with target DNA and attenuates the transcriptional activity. It increases the phosphorylation at Ser-256. Deacetylation by SIRT1 results in reactivation of the transcriptional activity. Oxidative stress by hydrogen peroxide treatment appears to promote deacetylation and uncoupling of insulin-induced phosphorylation. By contrast, resveratrol acts independently of acetylation.
Cytoplasm. Nucleus.
Note: Shuttles between the cytoplasm and nucleus. Largely nuclear in unstimulated cells. In osteoblasts, colocalizes with ATF4 and RUNX2 in the nucleus (By similarity). Insulin-induced phosphorylation at Ser-256 by PKB/AKT1 leads, via stimulation of Thr-24 phosphorylation, to binding of 14-3-3 proteins and nuclear export to the cytoplasm where it is degraded by the ubiquitin-proteosomal pathway. Phosphorylation at Ser-249 by CDK1 disrupts binding of 14-3-3 proteins and promotes nuclear accumulation. Phosphorylation by NLK results in nuclear export. Translocates to the nucleus upon oxidative stress-induced phosphorylation at Ser-212 by STK4/MST1. SGK1-mediated phosphorylation also results in nuclear translocation. Retained in the nucleus under stress stimuli including oxidative stress, nutrient deprivation or nitric oxide. Retained in the nucleus on methylation.
Ubiquitous.
Research Fields
· Cellular Processes > Cell growth and death > Cellular senescence. (View pathway)
· Environmental Information Processing > Signal transduction > FoxO signaling pathway. (View pathway)
· Environmental Information Processing > Signal transduction > AMPK signaling pathway. (View pathway)
· Human Diseases > Endocrine and metabolic diseases > Insulin resistance.
· Human Diseases > Infectious diseases: Viral > Human papillomavirus infection.
· Human Diseases > Cancers: Overview > Pathways in cancer. (View pathway)
· Human Diseases > Cancers: Overview > Transcriptional misregulation in cancer.
· Human Diseases > Cancers: Specific types > Prostate cancer. (View pathway)
· Organismal Systems > Aging > Longevity regulating pathway. (View pathway)
· Organismal Systems > Aging > Longevity regulating pathway - multiple species. (View pathway)
· Organismal Systems > Endocrine system > Insulin signaling pathway. (View pathway)
· Organismal Systems > Endocrine system > Thyroid hormone signaling pathway. (View pathway)
· Organismal Systems > Endocrine system > Glucagon signaling pathway.
References
Application: WB Species: Mouse Sample:
Application: WB Species: Rat Sample: H9c2 cells
Application: IHC Species: Rat Sample: H9c2 cells
Application: WB Species: Human Sample: HepG2 cells
Restrictive clause
Affinity Biosciences tests all products strictly. Citations are provided as a resource for additional applications that have not been validated by Affinity Biosciences. Please choose the appropriate format for each application and consult Materials and Methods sections for additional details about the use of any product in these publications.
For Research Use Only.
Not for use in diagnostic or therapeutic procedures. Not for resale. Not for distribution without written consent. Affinity Biosciences will not be held responsible for patent infringement or other violations that may occur with the use of our products. Affinity Biosciences, Affinity Biosciences Logo and all other trademarks are the property of Affinity Biosciences LTD.