APG5L/ATG5 Antibody - #DF6010
| Product: | APG5L/ATG5 Antibody |
| Catalog: | DF6010 |
| Description: | Rabbit polyclonal antibody to APG5L/ATG5 |
| Application: | WB IHC |
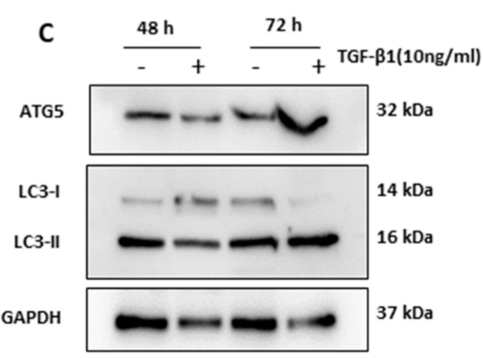
| Cited expt.: | WB |
| Reactivity: | Human, Mouse, Rat |
| Prediction: | Pig, Zebrafish, Bovine, Horse, Sheep, Rabbit, Dog, Chicken, Xenopus |
| Mol.Wt.: | 35kD,56kD(complex); 32kD(Calculated). |
| Uniprot: | Q9H1Y0 |
| RRID: | AB_2837987 |
Related Downloads
Protocols
Product Info
*The optimal dilutions should be determined by the end user. For optimal experimental results, antibody reuse is not recommended.
*Tips:
WB: For western blot detection of denatured protein samples. IHC: For immunohistochemical detection of paraffin sections (IHC-p) or frozen sections (IHC-f) of tissue samples. IF/ICC: For immunofluorescence detection of cell samples. ELISA(peptide): For ELISA detection of antigenic peptide.
Cite Format: Affinity Biosciences Cat# DF6010, RRID:AB_2837987.
Fold/Unfold
APG 5; APG 5L; APG5; APG5 autophagy 5 like; APG5 like; APG5-like; APG5L; Apoptosis specific protein; Apoptosis-specific protein; ASP; ATG 5; Atg5; ATG5 autophagy related 5 homolog; ATG5_HUMAN; Autophagy protein 5; Autophagy related 5; hAPG5; Homolog of S Cerevisiae autophagy 5; OTTHUMP00000040507;
Immunogens
A synthesized peptide derived from human APG5L/ATG5, corresponding to a region within C-terminal amino acids.
Ubiquitous. The mRNA is present at similar levels in viable and apoptotic cells, whereas the protein is dramatically highly expressed in apoptotic cells.
- Q9H1Y0 ATG5_HUMAN:
- Protein BLAST With
- NCBI/
- ExPASy/
- Uniprot
MTDDKDVLRDVWFGRIPTCFTLYQDEITEREAEPYYLLLPRVSYLTLVTDKVKKHFQKVMRQEDISEIWFEYEGTPLKWHYPIGLLFDLLASSSALPWNITVHFKSFPEKDLLHCPSKDAIEAHFMSCMKEADALKHKSQVINEMQKKDHKQLWMGLQNDRFDQFWAINRKLMEYPAEENGFRYIPFRIYQTTTERPFIQKLFRPVAADGQLHTLGDLLKEVCPSAIDPEDGEKKNQVMIHGIEPMLETPLQWLSEHLSYPDNFLHISIIPQPTD
Predictions
Score>80(red) has high confidence and is suggested to be used for WB detection. *The prediction model is mainly based on the alignment of immunogen sequences, the results are for reference only, not as the basis of quality assurance.
High(score>80) Medium(80>score>50) Low(score<50) No confidence
Research Backgrounds
Involved in autophagic vesicle formation. Conjugation with ATG12, through a ubiquitin-like conjugating system involving ATG7 as an E1-like activating enzyme and ATG10 as an E2-like conjugating enzyme, is essential for its function. The ATG12-ATG5 conjugate acts as an E3-like enzyme which is required for lipidation of ATG8 family proteins and their association to the vesicle membranes. Involved in mitochondrial quality control after oxidative damage, and in subsequent cellular longevity. Plays a critical role in multiple aspects of lymphocyte development and is essential for both B and T lymphocyte survival and proliferation. Required for optimal processing and presentation of antigens for MHC II. Involved in the maintenance of axon morphology and membrane structures, as well as in normal adipocyte differentiation. Promotes primary ciliogenesis through removal of OFD1 from centriolar satellites and degradation of IFT20 via the autophagic pathway.
May play an important role in the apoptotic process, possibly within the modified cytoskeleton. Its expression is a relatively late event in the apoptotic process, occurring downstream of caspase activity. Plays a crucial role in IFN-gamma-induced autophagic cell death by interacting with FADD.
(Microbial infection) May act as a proviral factor. In association with ATG12, negatively regulates the innate antiviral immune response by impairing the type I IFN production pathway upon vesicular stomatitis virus (VSV) infection. Required for the translation of incoming hepatitis C virus (HCV) RNA and, thereby, for initiation of HCV replication, but not required once infection is established.
Conjugated to ATG12; which is essential for autophagy, but is not required for association with isolation membrane.
Acetylated by EP300.
Cytoplasm. Preautophagosomal structure membrane>Peripheral membrane protein.
Note: Colocalizes with nonmuscle actin. The conjugate detaches from the membrane immediately before or after autophagosome formation is completed (By similarity). Localizes also to discrete punctae along the ciliary axoneme and to the base of the ciliary axoneme.
Ubiquitous. The mRNA is present at similar levels in viable and apoptotic cells, whereas the protein is dramatically highly expressed in apoptotic cells.
Belongs to the ATG5 family.
Research Fields
· Cellular Processes > Transport and catabolism > Autophagy - other. (View pathway)
· Cellular Processes > Transport and catabolism > Autophagy - animal. (View pathway)
· Cellular Processes > Cell growth and death > Ferroptosis. (View pathway)
· Human Diseases > Infectious diseases: Bacterial > Shigellosis.
· Organismal Systems > Aging > Longevity regulating pathway. (View pathway)
· Organismal Systems > Aging > Longevity regulating pathway - multiple species. (View pathway)
· Organismal Systems > Immune system > NOD-like receptor signaling pathway. (View pathway)
· Organismal Systems > Immune system > RIG-I-like receptor signaling pathway. (View pathway)
References
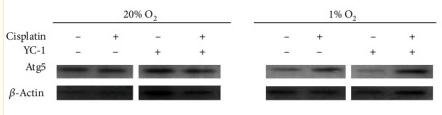
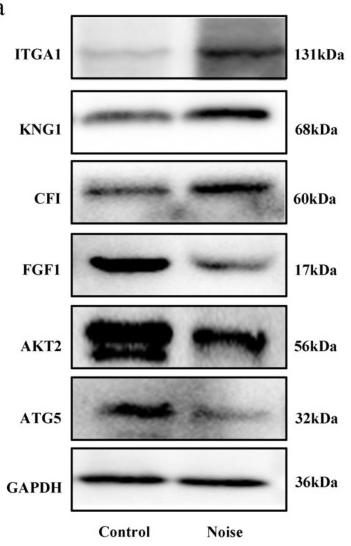
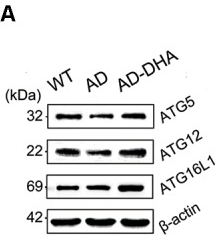
Application: WB Species: Mouse Sample: brain tissue
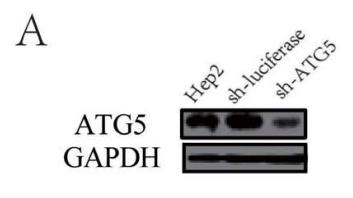
Application: WB Species: human Sample: Hep2 cells
Restrictive clause
Affinity Biosciences tests all products strictly. Citations are provided as a resource for additional applications that have not been validated by Affinity Biosciences. Please choose the appropriate format for each application and consult Materials and Methods sections for additional details about the use of any product in these publications.
For Research Use Only.
Not for use in diagnostic or therapeutic procedures. Not for resale. Not for distribution without written consent. Affinity Biosciences will not be held responsible for patent infringement or other violations that may occur with the use of our products. Affinity Biosciences, Affinity Biosciences Logo and all other trademarks are the property of Affinity Biosciences LTD.