Cytochrome P450 7A1 Antibody - #DF2612
| Product: | Cytochrome P450 7A1 Antibody |
| Catalog: | DF2612 |
| Description: | Rabbit polyclonal antibody to Cytochrome P450 7A1 |
| Application: | WB IHC IF/ICC |
| Cited expt.: | WB |
| Reactivity: | Human, Mouse, Rat |
| Prediction: | Pig, Horse, Rabbit, Dog |
| Mol.Wt.: | 58 kDa; 58kD(Calculated). |
| Uniprot: | P22680 |
| RRID: | AB_2839818 |
Product Info
*The optimal dilutions should be determined by the end user. For optimal experimental results, antibody reuse is not recommended.
*Tips:
WB: For western blot detection of denatured protein samples. IHC: For immunohistochemical detection of paraffin sections (IHC-p) or frozen sections (IHC-f) of tissue samples. IF/ICC: For immunofluorescence detection of cell samples. ELISA(peptide): For ELISA detection of antigenic peptide.
Cite Format: Affinity Biosciences Cat# DF2612, RRID:AB_2839818.
Fold/Unfold
Cholesterol 7 alpha hydroxylase; Cholesterol 7 alpha monooxygenase; Cholesterol 7-alpha-hydroxylase; Cholesterol 7-alpha-monooxygenase; CP7A; CP7A1_HUMAN; CYP 7; CYP7; CYP7A1; CYPVII; Cytochrome P450 7A1; Cytochrome P450, family 7, subfamily A, polypeptide 1; Cytochrome P450, subfamily VIIA (cholesterol 7 alpha monooxygenase),; Cytochrome P450, subfamily VIIA (cholesterol 7 alpha-monooxygenase), polypeptide 1; MGC126826; MGC138389;
Immunogens
A synthesized peptide derived from human Cytochrome P450 7A1, corresponding to a region within the internal amino acids.
- P22680 CP7A1_HUMAN:
- Protein BLAST With
- NCBI/
- ExPASy/
- Uniprot
MMTTSLIWGIAIAACCCLWLILGIRRRQTGEPPLENGLIPYLGCALQFGANPLEFLRANQRKHGHVFTCKLMGKYVHFITNPLSYHKVLCHGKYFDWKKFHFATSAKAFGHRSIDPMDGNTTENINDTFIKTLQGHALNSLTESMMENLQRIMRPPVSSNSKTAAWVTEGMYSFCYRVMFEAGYLTIFGRDLTRRDTQKAHILNNLDNFKQFDKVFPALVAGLPIHMFRTAHNAREKLAESLRHENLQKRESISELISLRMFLNDTLSTFDDLEKAKTHLVVLWASQANTIPATFWSLFQMIRNPEAMKAATEEVKRTLENAGQKVSLEGNPICLSQAELNDLPVLDSIIKESLRLSSASLNIRTAKEDFTLHLEDGSYNIRKDDIIALYPQLMHLDPEIYPDPLTFKYDRYLDENGKTKTTFYCNGLKLKYYYMPFGSGATICPGRLFAIHEIKQFLILMLSYFELELIEGQAKCPPLDQSRAGLGILPPLNDIEFKYKFKHL
Predictions
Score>80(red) has high confidence and is suggested to be used for WB detection. *The prediction model is mainly based on the alignment of immunogen sequences, the results are for reference only, not as the basis of quality assurance.
High(score>80) Medium(80>score>50) Low(score<50) No confidence
Research Backgrounds
A cytochrome P450 monooxygenase involved in the metabolism of endogenous cholesterol and its oxygenated derivatives (oxysterols). Mechanistically, uses molecular oxygen inserting one oxygen atom into a substrate, and reducing the second into a water molecule, with two electrons provided by NADPH via cytochrome P450 reductase (CPR; NADPH-ferrihemoprotein reductase). Functions as a critical regulatory enzyme of bile acid biosynthesis and cholesterol homeostasis. Catalyzes the hydroxylation of carbon hydrogen bond at 7-alpha position of cholesterol, a rate-limiting step in cholesterol catabolism and bile acid biosynthesis. 7-alpha hydroxylates several oxysterols, including 4beta-hydroxycholesterol and 24-hydroxycholesterol. Catalyzes the oxidation of the 7,8 double bond of 7-dehydrocholesterol and lathosterol with direct and predominant formation of the 7-keto derivatives.
Endoplasmic reticulum membrane>Single-pass membrane protein. Microsome membrane>Single-pass membrane protein.
Detected in liver.
Belongs to the cytochrome P450 family.
Research Fields
· Metabolism > Lipid metabolism > Primary bile acid biosynthesis.
· Metabolism > Lipid metabolism > Steroid hormone biosynthesis.
· Metabolism > Global and overview maps > Metabolic pathways.
· Organismal Systems > Endocrine system > PPAR signaling pathway.
· Organismal Systems > Digestive system > Cholesterol metabolism.
References
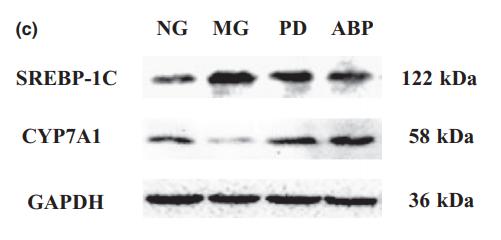
Application: WB Species: Mouse Sample: liver
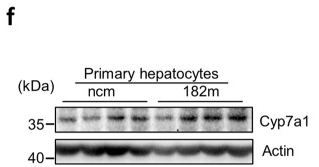
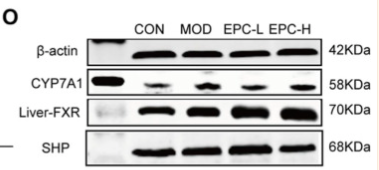
Application: WB Species: Rat Sample:
Restrictive clause
Affinity Biosciences tests all products strictly. Citations are provided as a resource for additional applications that have not been validated by Affinity Biosciences. Please choose the appropriate format for each application and consult Materials and Methods sections for additional details about the use of any product in these publications.
For Research Use Only.
Not for use in diagnostic or therapeutic procedures. Not for resale. Not for distribution without written consent. Affinity Biosciences will not be held responsible for patent infringement or other violations that may occur with the use of our products. Affinity Biosciences, Affinity Biosciences Logo and all other trademarks are the property of Affinity Biosciences LTD.