Integrin alpha V Antibody - #AF5152
| Product: | Integrin alpha V Antibody |
| Catalog: | AF5152 |
| Description: | Rabbit polyclonal antibody to Integrin alpha V |
| Application: | WB IHC |
| Cited expt.: | WB |
| Reactivity: | Human, Mouse |
| Prediction: | Pig, Bovine, Horse, Sheep, Rabbit, Dog |
| Mol.Wt.: | 116 kDa; 116kD(Calculated). |
| Uniprot: | P06756 |
| RRID: | AB_2837638 |
Related Downloads
Protocols
Product Info
*The optimal dilutions should be determined by the end user. For optimal experimental results, antibody reuse is not recommended.
*Tips:
WB: For western blot detection of denatured protein samples. IHC: For immunohistochemical detection of paraffin sections (IHC-p) or frozen sections (IHC-f) of tissue samples. IF/ICC: For immunofluorescence detection of cell samples. ELISA(peptide): For ELISA detection of antigenic peptide.
Cite Format: Affinity Biosciences Cat# AF5152, RRID:AB_2837638.
Fold/Unfold
antigen identified by monoclonal antibody L230; CD 51; CD51; DKFZp686A08142; integrin alpha V beta 3; integrin alpha-V; Integrin alpha-V light chain; integrin alphaVbeta3; integrin, alpha V (vitronectin receptor, alpha polypeptide, antigen CD51); ITAV_HUMAN; ITGAV; MSK 8; Msk8; Vitronectin receptor subunit alpha; VNRA; VTNR;
Immunogens
A synthesized peptide derived from human Integrin alpha V, corresponding to a region within the internal amino acids.
- P06756 ITAV_HUMAN:
- Protein BLAST With
- NCBI/
- ExPASy/
- Uniprot
MAFPPRRRLRLGPRGLPLLLSGLLLPLCRAFNLDVDSPAEYSGPEGSYFGFAVDFFVPSASSRMFLLVGAPKANTTQPGIVEGGQVLKCDWSSTRRCQPIEFDATGNRDYAKDDPLEFKSHQWFGASVRSKQDKILACAPLYHWRTEMKQEREPVGTCFLQDGTKTVEYAPCRSQDIDADGQGFCQGGFSIDFTKADRVLLGGPGSFYWQGQLISDQVAEIVSKYDPNVYSIKYNNQLATRTAQAIFDDSYLGYSVAVGDFNGDGIDDFVSGVPRAARTLGMVYIYDGKNMSSLYNFTGEQMAAYFGFSVAATDINGDDYADVFIGAPLFMDRGSDGKLQEVGQVSVSLQRASGDFQTTKLNGFEVFARFGSAIAPLGDLDQDGFNDIAIAAPYGGEDKKGIVYIFNGRSTGLNAVPSQILEGQWAARSMPPSFGYSMKGATDIDKNGYPDLIVGAFGVDRAILYRARPVITVNAGLEVYPSILNQDNKTCSLPGTALKVSCFNVRFCLKADGKGVLPRKLNFQVELLLDKLKQKGAIRRALFLYSRSPSHSKNMTISRGGLMQCEELIAYLRDESEFRDKLTPITIFMEYRLDYRTAADTTGLQPILNQFTPANISRQAHILLDCGEDNVCKPKLEVSVDSDQKKIYIGDDNPLTLIVKAQNQGEGAYEAELIVSIPLQADFIGVVRNNEALARLSCAFKTENQTRQVVCDLGNPMKAGTQLLAGLRFSVHQQSEMDTSVKFDLQIQSSNLFDKVSPVVSHKVDLAVLAAVEIRGVSSPDHVFLPIPNWEHKENPETEEDVGPVVQHIYELRNNGPSSFSKAMLHLQWPYKYNNNTLLYILHYDIDGPMNCTSDMEINPLRIKISSLQTTEKNDTVAGQGERDHLITKRDLALSEGDIHTLGCGVAQCLKIVCQVGRLDRGKSAILYVKSLLWTETFMNKENQNHSYSLKSSASFNVIEFPYKNLPIEDITNSTLVTTNVTWGIQPAPMPVPVWVIILAVLAGLLLLAVLVFVMYRMGFFKRVRPPQEEQEREQLQPHENGEGNSET
Predictions
Score>80(red) has high confidence and is suggested to be used for WB detection. *The prediction model is mainly based on the alignment of immunogen sequences, the results are for reference only, not as the basis of quality assurance.
High(score>80) Medium(80>score>50) Low(score<50) No confidence
Research Backgrounds
The alpha-V (ITGAV) integrins are receptors for vitronectin, cytotactin, fibronectin, fibrinogen, laminin, matrix metalloproteinase-2, osteopontin, osteomodulin, prothrombin, thrombospondin and vWF. They recognize the sequence R-G-D in a wide array of ligands. ITGAV:ITGB3 binds to fractalkine (CX3CL1) and may act as its coreceptor in CX3CR1-dependent fractalkine signaling. ITGAV:ITGB3 binds to NRG1 (via EGF domain) and this binding is essential for NRG1-ERBB signaling. ITGAV:ITGB3 binds to FGF1 and this binding is essential for FGF1 signaling. ITGAV:ITGB3 binds to FGF2 and this binding is essential for FGF2 signaling. ITGAV:ITGB3 binds to IGF1 and this binding is essential for IGF1 signaling. ITGAV:ITGB3 binds to IGF2 and this binding is essential for IGF2 signaling. ITGAV:ITGB3 binds to IL1B and this binding is essential for IL1B signaling. ITGAV:ITGB3 binds to PLA2G2A via a site (site 2) which is distinct from the classical ligand-binding site (site 1) and this induces integrin conformational changes and enhanced ligand binding to site 1. ITGAV:ITGB3 and ITGAV:ITGB6 act as a receptor for fibrillin-1 (FBN1) and mediate R-G-D-dependent cell adhesion to FBN1. Integrin alpha-V/beta-6 or alpha-V/beta-8 (ITGAV:ITGB6 or ITGAV:ITGB8) mediates R-G-D-dependent release of transforming growth factor beta-1 (TGF-beta-1) from regulatory Latency-associated peptide (LAP), thereby playing a key role in TGF-beta-1 activation. ITGAV:ITGB3 act as a receptor for CD40LG.
(Microbial infection) Integrin ITGAV:ITGB5 acts as a receptor for Adenovirus type C.
(Microbial infection) Integrin ITGAV:ITGB5 and ITGAV:ITGB3 act as receptors for Coxsackievirus A9 and B1.
(Microbial infection) Integrin ITGAV:ITGB3 acts as a receptor for Herpes virus 8/HHV-8.
(Microbial infection) Integrin ITGAV:ITGB6 acts as a receptor for herpes simplex 1/HHV-1.
(Microbial infection) Integrin ITGAV:ITGB3 acts as a receptor for Human parechovirus 1.
(Microbial infection) Integrin ITGAV:ITGB3 acts as a receptor for West nile virus.
(Microbial infection) In case of HIV-1 infection, the interaction with extracellular viral Tat protein seems to enhance angiogenesis in Kaposi's sarcoma lesions.
Cell membrane>Single-pass type I membrane protein. Cell junction>Focal adhesion.
Belongs to the integrin alpha chain family.
Research Fields
· Cellular Processes > Transport and catabolism > Phagosome. (View pathway)
· Cellular Processes > Cellular community - eukaryotes > Focal adhesion. (View pathway)
· Cellular Processes > Cell motility > Regulation of actin cytoskeleton. (View pathway)
· Environmental Information Processing > Signal transduction > PI3K-Akt signaling pathway. (View pathway)
· Environmental Information Processing > Signaling molecules and interaction > ECM-receptor interaction. (View pathway)
· Environmental Information Processing > Signaling molecules and interaction > Cell adhesion molecules (CAMs). (View pathway)
· Human Diseases > Infectious diseases: Viral > Human papillomavirus infection.
· Human Diseases > Cancers: Overview > Pathways in cancer. (View pathway)
· Human Diseases > Cancers: Overview > Proteoglycans in cancer.
· Human Diseases > Cancers: Specific types > Small cell lung cancer. (View pathway)
· Human Diseases > Cardiovascular diseases > Hypertrophic cardiomyopathy (HCM).
· Human Diseases > Cardiovascular diseases > Arrhythmogenic right ventricular cardiomyopathy (ARVC).
· Human Diseases > Cardiovascular diseases > Dilated cardiomyopathy (DCM).
· Organismal Systems > Endocrine system > Thyroid hormone signaling pathway. (View pathway)
References
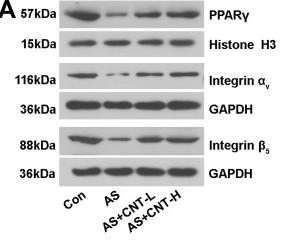
Application: WB Species: Mice Sample: aortic tissues and RAW264.7 cells
Restrictive clause
Affinity Biosciences tests all products strictly. Citations are provided as a resource for additional applications that have not been validated by Affinity Biosciences. Please choose the appropriate format for each application and consult Materials and Methods sections for additional details about the use of any product in these publications.
For Research Use Only.
Not for use in diagnostic or therapeutic procedures. Not for resale. Not for distribution without written consent. Affinity Biosciences will not be held responsible for patent infringement or other violations that may occur with the use of our products. Affinity Biosciences, Affinity Biosciences Logo and all other trademarks are the property of Affinity Biosciences LTD.